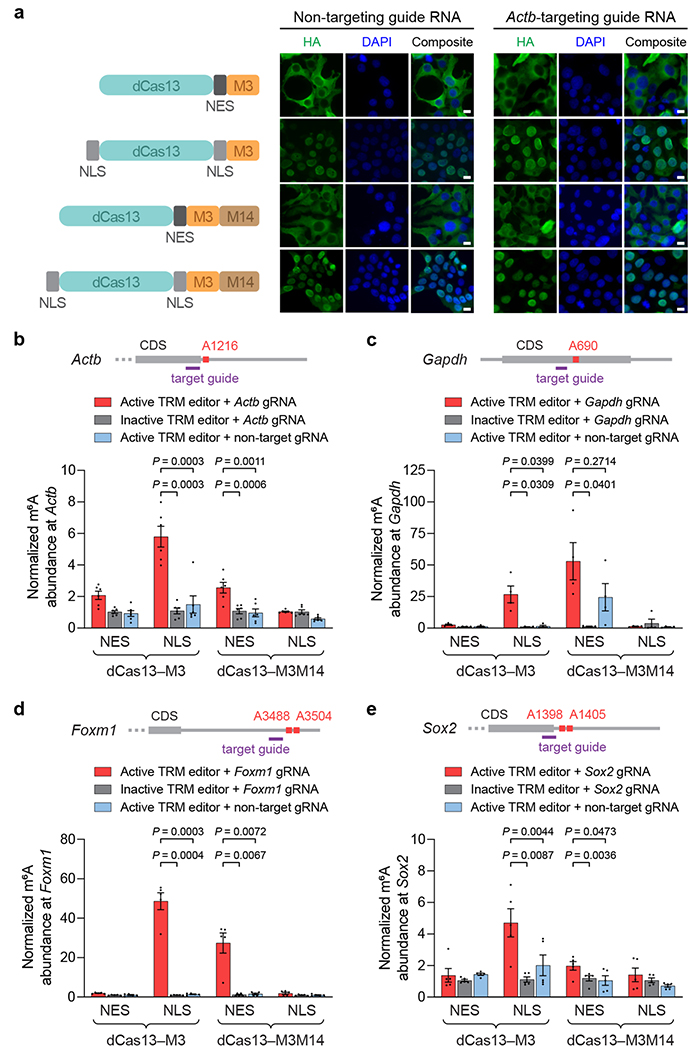
Figure 4. Cellular localization of TRM editors and targeted methylation of endogenous transcripts in human cells.
(a) Left: nucleus- and cytoplasm-localized TRM editor variants. NES = nuclear export signal; NLS = nuclear localization signal. Right: representative immunofluorescence images of HEK293T cells transfected with HA-tagged TRM editors and non-targeting or Actb-targeting guide RNAs. Green = HA tag; blue = DAPI. Scale bars represent 10 μm. The experiment was independently performed twice with similar results. (b-e) Methylation by nucleus- and cytoplasm-localized TRM editors targeting endogenous (b) Actb A1216, (c) Gapdh A690, (d) Foxm1 A3488 and A3504, and (e) Sox2 A1398 and A1405. Guide RNAs used for targeting each transcript are shown in purple. Inactive TRM editors contain a methyltransferase-inactivating D395A mutation within M3. Values and error bars reflect the mean±s.e.m. of n=6 (b), n=4 (c), or n=5 (d,e) independent biological replicates. P values were calculated using a two-tailed Student’s t-test.