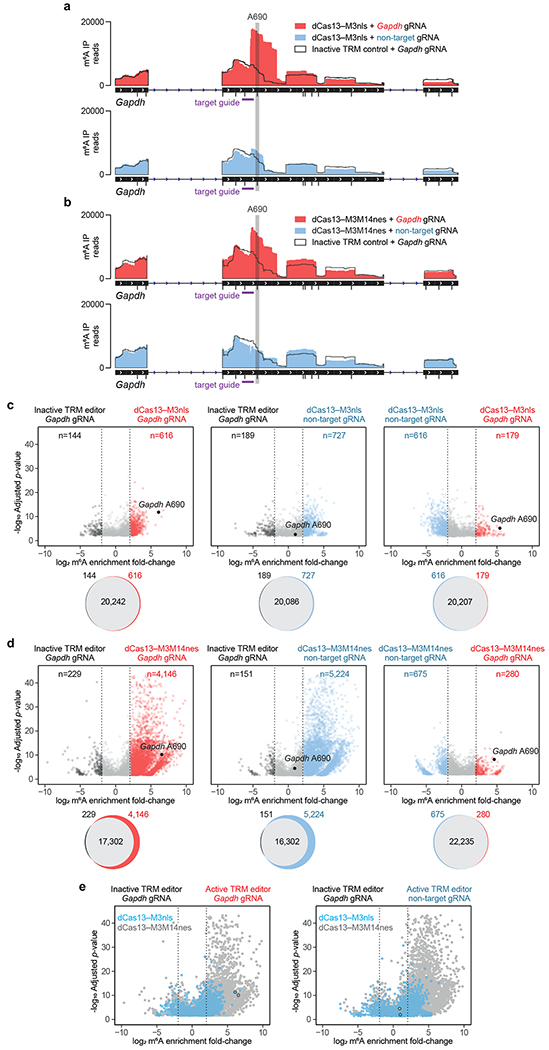
Figure 5. Specificity and off-target methylation of TRM editors.
(a,b) Gapdh read coverage of m6A-immunoprecipitated (m6A IP) RNA from MeRIP-seq of transfected HEK293T cells. Gapdh A690 was targeted with (a) dCas13–M3nls or (b) dCas13–M3M14nes with the following conditions: active editor with a target guide RNA, inactive editor with a target guide RNA, and active editor with a non-target guide RNA. All DRACH motifs susceptible to TRM modification are shown as black tick marks underneath the IGV track. The target guide RNA (purple) and targeted A690 site (grey) are shown. (c,d) Differential m6A enrichment of >21,000 methylated sites in HEK293T cells transfected with (c) dCas13–M3nls or (d) dCas13–M3M14nes and Gapdh A690-targeting or non-target guide RNAs. Top: differential methylation of m6A sites between conditions indicated above. Only differentially methylated sites with statistical significance (P<0.001) are shown. Bottom: Venn diagrams depicting overlap of all methylated m6A sites for the above comparisons. (e) Overlay of dCas13–M3nls (blue) and dCas13–M3M14nes (grey) differential methylation with the following comparisons: left, active editors with Gapdh guide RNAs vs. inactive editors with Gapdh guide RNAs; right, active editors with non-target guide RNAs vs. inactive editors with Gapdh guide RNAs. The targeted Gapdh A690 site is outlined in black. Inactive TRM editors contain a methyltransferase-inactivating D395A mutation within M3. MeRIP-seq analysis was performed with n=5 independent biological replicates. Statistical significance was calculated using a logs likelihood-ratio test with false discovery rate correction.