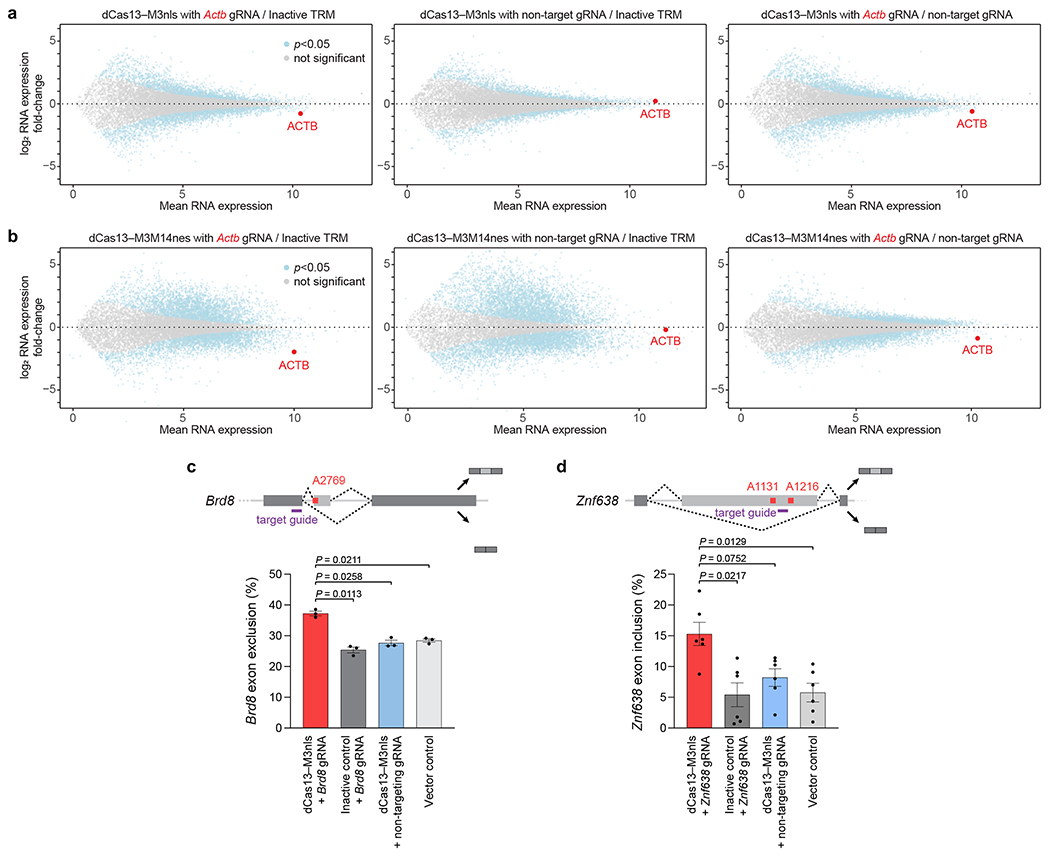
Figure 6. TRM effect on RNA abundance and alternative splicing.
Differential RNA expression of HEK293T cells transfected with (a) dCas13–M3nls or (b) dCas13–M3M14nes and either Actb A1216-targeting or non-targeting guide RNAs. The targeted Actb gene is marked in red. Inactive TRM control indicates editors containing a methyltransferase-inactivating M3 D395A mutation with an Actb-targeting guide RNA. Non-target gRNA indicates methyltransferase-active TRM editors with a non-targeting guide RNA as a control. Differential RNA-seq analysis was performed with n=5 independent biological replicates. Statistical significance was calculated using a two-tailed Student’s t-test with false discovery rate correction. Alternative splicing of (c) Brd8 and (d) Znf638 in HEK293T cells transfected with dCas13–M3nls and the indicated guide RNAs (purple) for targeting m6A sites shown (red). Exon exclusion or inclusion indicates the percentage of RNA isoform lacking or containing the alternatively-spliced exon, respectively. Values and error bars reflect the mean±s.e.m. of n=3 (c) or n=6 (d) independent biological replicates. P values were calculated using a two-tailed Student’s t-test.