Figure 7. CDDO-Im inhibits interferon stimulated gene expression in RAW264.7 cells.
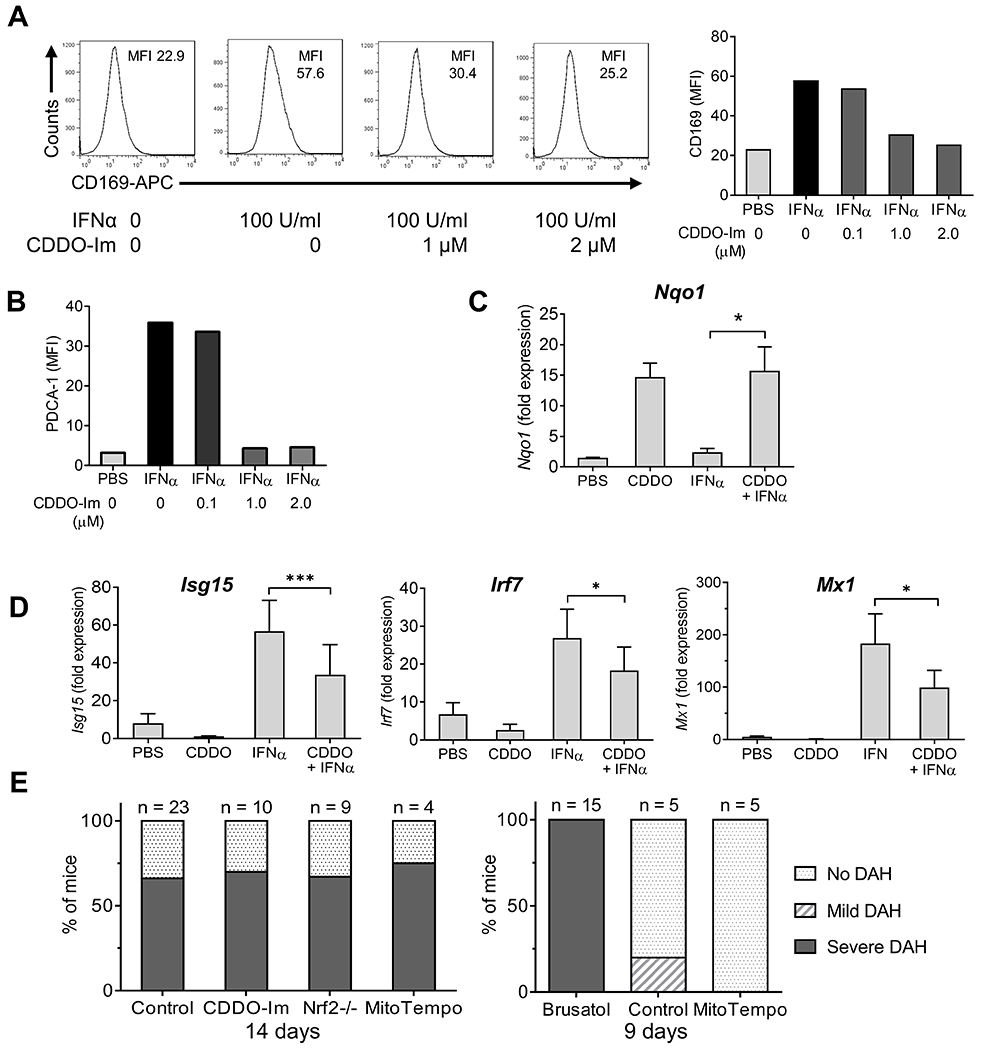
A, RAW264.7 cells were pretreated with CDDO-Im (0.1, 1.0, or 2.0 μM in DMSO) or vehicle alone for 1-h, and then challenged with IFNα4 (100 U/ml) for 24-h. Surface staining of the IFN-I regulated protein CD169 (MFI) was analyzed by flow cytometry. Left, representative histograms of CD169 surface staining; right, quantification. B, Quantification of surface staining (flow cytometry) for a second IFN-inducible protein (PDCA-1) showing a pattern similar to CD169. C-D, RAW264.7 cells were pretreated with CDDO-Im (0.5 μM) or PBC and then challenged with IFNα4 (100 U/ml in PBS) or PBS alone for 6-h. C, Nrf2-regulated gene expression (Nqo1) relative to 18S RNA (qPCR) was determined (qPCR). D, ISG expression (Isg15, Irf7, Mx1) relative to 18S RNA (qPCR) was determined (qPCR). E, Mice were injected with pristane plus CDDO-Im (2.5 mg/kg in PBS i.p. every other day), brusatol (2 mg/kg in PBS i.p. every other day), MitoTEMPO (1.5 mg/kg in PBS i.p.), or vehicle. Left, DAH was assessed at 14-d in B6 mice receiving CDDO-Im, MitoTEMPO, or vehicle (Control) and in B6 Nrf2−/− mice. Right, DAH was assessed at 9-d in B6 mice receiving brusatol, MitoTEMPO, or vehicle. * P < 0.05, ** P < 0.01, *** P < 0.001 (Student t-test).
