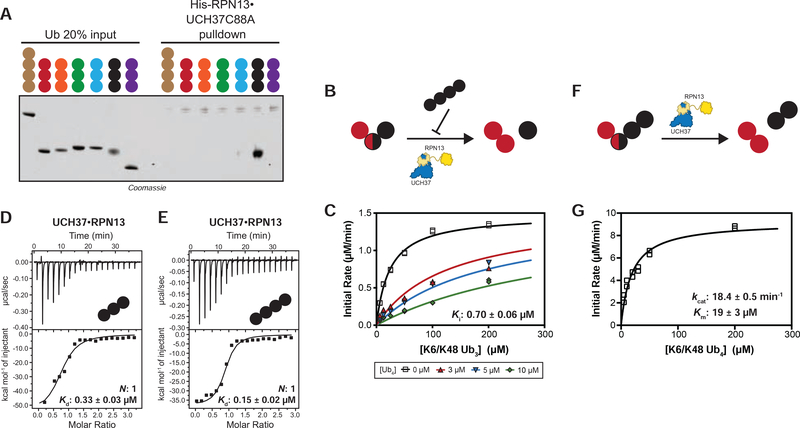
FIGURE 4. UCH37 Preferentially Binds K48 Chains.
(A) SDS-PAGE analysis of pull-downs with M1-, K6-, K11-, K29-, K33-, K48-, and K63-linked Ub chains. Each chain (50 pmol) was captured using immobilized His-RPN13•UCH37 C88A (5 nmol).
(B) Schematic for measuring the hydrolysis of native K6/K48 branched tri-Ub in the presence of K48 tetra-Ub added in trans.
(C) Michaelis-Menten plot for the hydrolysis of native K6/K48 branched tri-Ub by UCH37•RPN13 (0.5 μM) in the presence of K48 tetra-Ub.
(D-E) ITC analysis of active UCH37•RPN13 binding to K48-linked tri-Ub (C) and tetra-Ub (D).
(F) Schematic for measuring the hydrolysis of native K6/K48 branched tetra-Ub in cis.
(G) Michaelis-Menten plot for the hydrolysis of native K6/K48 branched tetra-Ub by UCH37•RPN13 (0.5 μM).
All ITC curves are representative traces and reported Kds (B, C) are derived from averaging fits of 2 independent experiments. All kinetic curves (A, D) are representative traces and constants derived from averaging fits of 2 independent experiments with SD. See also Figure S4 and Table S1D.