Abstract
Background
Transcriptional regulators are seminal players in the onset and progression of prostate cancer. However, clarification of their underlying regulatory circuits and mechanisms demands considerable effort.
Methods
Integrated analyses were performed on genomic, transcriptomic, and clinicopathological profiles of primary prostate cancer and transcription factor-binding profiles, which included estimating transcription factor activity, identifying transcription factors of prognostic values, and discovering cis- and trans-regulations by long noncoding RNAs. Interactions between transcription factors and long noncoding RNAs were validated by RNA immunoprecipitation quantitative PCR. RNA interference assays were performed to explore roles of the selected transcription regulators.
Findings
Sixteen transcription factors, namely, ETS1, ARID4B, KLF12, GMEB1, HBP1, MXI1, MYC, MAX, PGR, BCL11A, AR, KLF4, SRF, HIF1A, EHF, and ATOH1, were jointly identified as a prognostic signature. Candidate long noncoding RNAs interplaying with the prognostic signature constituent transcription factors were further discovered. Their interactions were randomly checked, and many of them were experimentally proved. Transcription regulation by MYC and its long noncoding RNA partner AL590617.2 was further validated on their candidate targets. Moreover, the regulatory network governed by the transcription factors and their interacting long noncoding RNA partners is illustrated and stored in our LNCTRN database (https://navy.shinyapps.io/lnctrn).
Interpretation
The prognostic signature constituent transcription factors and their interacting long noncoding RNAs may represent promising biomarkers and/or therapeutic targets for prostate cancer. Furthermore, the computational framework proposed in the present study can be utilized to explore critical transcriptional regulators in other types of cancer.
Funding
This work was supported by National Natural Science Foundation of China and Fudan University.
Keywords: Transcription factor, Long noncoding RNA, Interaction partner, Transcriptional network, Prostate cancer
Research in context.
Evidence before this study
Considerable advances illuminate the importance of transcriptional dysregulation in cancer including prostate cancer as a driving event that provokes the acquisition of cancer hallmarks. Clarification of the underlying transcriptional networks and mechanisms still demands a lot of effort.
Added value of this study
In the present study, we reveal the transcriptional network governed by the prognostic signature transcription factors and their interacting long noncoding RNAs in primary prostate cancer, which can be accessed via our LNCTRN database (https://navy.shinyapps.io/lnctrn).
Implications of all the available evidence
The prognostic signature transcription factors and their interacting long noncoding RNAs may represent potential biomarkers and/or therapeutic targets in prostate cancer. In addition, to the best of our knowledge, the present study is the first to recognize the interaction between transcription factors and long noncoding RNAs in a large scale. The computational framework proposed in our study can be used to discover critical transcriptional regulators in other cancer types.
Alt-text: Unlabelled box
1. Introduction
Considerable advances illuminate the importance of transcriptional dysregulation in cancer including prostate cancer as a driving event that provokes the acquisition of cancer hallmarks [1,2]. A genome-wide association study discovered 77 prostate cancer risk loci the majority of which overlap putative enhancers [3]. Two single nucleotide variants were further linked to the androgen sensitivity of enhancers in prostate cancer cells through affecting the binding of androgen receptor.
Androgen receptor (AR) acts as a transcription factor and mediates the cellular action of androgen, which is required for development, maintenance and function of the prostate gland [4]. The same as normal prostate cells, proliferation and survival of prostate cancer cells highly rely on androgen and AR, providing the rationale for androgen deprivation therapy in management of prostate cancer [1,2,5]. Deprivation is typically achieved by oral administration of androgen antagonist to disrupt the binding of androgen to AR and thereby suppress the androgen signaling axis. However, progression of prostate cancer to castration resistance is inevitable [6]. A leading cause for resistance is AR gene amplification and aberrant activation that restore the androgen signaling and allow the cancer cell growth despite a low level of circulating androgen [1,5,6]. More potent agents targeting AR have been developed, and some of them such as enzalutamide have shown promising clinical benefit in patients with castration-resistant prostate cancer [1,2,5,6]. The antiandrogen enzalutamide not only competes with androgen for AR binding but also prevent AR translocation to nucleus, and was recently approved for castration-resistant prostate cancer.
Several other transcription factors have been implicated in the prostate cancer as well. Translocation of ERG, a member of ETS family, to downstream of androgen-dependent TMPRSS2 promoter was observed in approximate half of prostate tumors, which results in elevated expression of ERG [7]. The rearrangement is an early event in prostate cancer and drives the initiation of prostate cancer [8]. ERG overexpression leads to global chromatin reorganization [9] and promotes cell invasion and neoplastic transformation [8]. Roles of MYC in the tumorigenesis have been the subjects of intense investigation for decades [10], [11], [12]. MYC overexpression contributes to multiple hallmarks of cancer. Recurrent overexpression of MYC protein has been detected at early stages of prostate cancer and is believed to drive the prostate carcinogenesis [11,12]. In addition, MYC is a central driving force of the evolution of prostate cancer to an androgen-independent state.
Identifying and characterizing long noncoding RNAs (lncRNAs) is undoubtedly a burgeoning field of the transcriptional regulation research. Nuclear lncRNAs can change chromatin organization by interacting with chromatin modulators [13,14]. NEAT1, a poor prognostic factor tied up with the metastatic recurrence and castration resistance of prostate cancer, has demonstrated the ability to modulate the chromatin status evidenced by its direction interaction with active histone H3 modifications at target promoters upon NEAT1 overexpression, thus leading to upregulation of oncogenic genes independent of AR [15]. ANRIL mediates epigenetic repression of the INK4 cell cycle inhibitors through recruiting the polycomb repressive complex 1 to the INK4 gene locus, which explains elevated level of ANRIL in the prostate cancer [16]. In addition, it is reported that lncRNAs activate or repress transcription through either recruiting transcription factors to or sequestering them from regulatory regions [13,14]. Cooperative binding of the lncRNAs PCGEM1 and PRNCR1 to AR further recruits PYGO2, which selectively reinforces the looping of AR bound enhancers over target gene promoters even in the absence of androgen and thereby promotes resistance of prostate cancer cells to the androgen deprivation [17]. Notably, the interaction of AR, PCGEM1 and PRNCR1 is specific to the prostate cancer and has not been detected in the normal prostate. However, a recent publication refuted the interaction between AR and those two lncRNAs in the prostate cancer [18]. Thus, the interaction of AR, PCGEM1 and PRNCR1 needs more delicate investigation. LncRNAs also impact localization, stability and activity of transcription factors via direct physical interaction [13,14]. HOTAIR binding to the AR protein protects AR from degradation by blocking the interplay between AR and a E3 ubiquitin ligase MDM2 and therefore preventing the AR ubiquitination [19]. HOTAIR overexpression in prostate cancer stimulates the androgen-independent AR activity and finally drives castration resistance.
It is notable that lncRNAs generally exhibit a cell- and/or tissue/tumor-specific expression pattern, making them attractive candidates for diagnostic markers or therapeutics [20]. Testing for PCA3, the first approved lncRNA biomarker in cancer, has greatly improved the specificity of prostate cancer detection [21]. Rapid evolution and increased success of the nucleic acid-based clinical trials have encouraged more preclinical research into lncRNAs [20].
In the present study, integrated analyses were performed on genomic, transcriptomic and clinicopathological profiles of the primary prostate cancers from the Cancer Genome Atlas (TCGA, https://portal.gdc.cancer.gov) and transcription factor binding sites from the GTRD ChIP-Seq database [22]. 16 transcription factors were identified to work jointly as a prognostic signature for the prostate cancer. In addition, transcriptional regulation by long noncoding RNAs were predicted and deposited in our LNCTRN database (https://navy.shinyapps.io/lnctrn). Furthermore, our study was focused on the 16 prognostic transcription factors and their interacting lncRNA partners. The interactions were randomly validated by RNA immunoprecipitation, the majority of which were proved.
2. Materials and methods
2.1. Data accession and processing
The copy number variation, genomic mutation, gene expression, and clinicopathological profiles of primary prostate cancer were all downloaded from the GDC data portal of the Cancer Genome Atlas (TCGA, https://portal.gdc.cancer.gov) which covered 498 primary tumors and 52 normal tissues. A total of 20433 genes with RNA-Seq counts per million values > 0.5 in over 10% of the examined samples were considered in our analysis. Subsequently, gene expression estimation and differential gene expression analysis were performed on the RNA-Seq counts using edgeR [23]. In the former task, fragments per kilobase per million mapped fragments (FPKM) values were calculated, log2 transformed, and then finally Z-score scaled. The standardized values were called zFPKM, which estimated the relative gene expression levels across samples. A total of 5946 genes were differentially expressed between tumors and normal tissues (fold change > 1.5 and false discovery rate [FDR] < 0.05). In addition, molecular subtypes, preoperative PSA levels, and tumor cellularity were obtained through TCGAbiolinks [24].
From the GTRD ChIP-Seq database [22], we extracted transcription factor-binding sites in all collected cell types, which were detected by at least three peak calling algorithms. A gene was considered as the target of a transcription factor if the transcription factor bound to its proximal regulatory region. Herein, proximal regulatory regions referred to genomic regions within 1 kb from gene transcription start sites.
2.2. Estimation of transcription factor activity
In total, 5946 differentially expressed genes were used in the calculation of transcription factor activity. Multiple linear regression was applied to estimate transcription factor activity in each sample as follows:
where Eti is the zFPKM value of the ith gene in the tth sample, xij is number of binding sites of the jth transcription factor within 1 kb from the transcription start sites of the ith gene, βtj estimates the activity of the jth transcription factor in the tth sample, βt0 is the constant term in the model, and εti is the random error for the ith gene in the tth sample. Transcription factors without statistical significance (p-value > 0.01) were removed from the regression model.
Finally, transcription factors active in less than 5% of the samples were not included in further analyses. Limma [25] was used to analyze differences between cancerous and normal prostate tissues in transcription factor activity (FDR < 0.05).
2.3. Prediction of transcriptional regulation by lncRNAs
Similarly, 5946 differentially expressed genes were used in the prediction of transcriptional regulation by lncRNAs. First, expression correlation between each of these genes and each lncRNA was measured by Pearson correlation of their zFPKM values.
An lncRNA was considered to transcriptionally regulate a gene in cis, if it satisfied two criteria: (1) their expression correlation was high (|coefficient| > 0.3 and FDR < 0.01) and (2) the minimum distance between their transcription start sites was smaller than 1 kb.
An lncRNA was considered to regulate a target in trans by forming an RNA–DNA triplex in the proximal regulatory region if Triplexator [26] detected that the lncRNA bound a DNA fragment within 1 kb from the target transcription start sites. Triplexator was implemented with default parameters, except that the minimum length of a putative triplex forming oligonucleotide, triplex target site, or triplex was set as 20 bp. The minimum distance between the transcription start sites of the lncRNA and its target gene must also be farther than 1 kb. In addition, expression correlation was considered (FDR < 0.01).
To predict the interaction between a transcription factor and an lncRNA, we narrowed down the differentially expressed genes to the transcription factor targets and then measured Spearman correlation between their expression correlations (Pearson correlation coefficients) with the lncRNA and binding site numbers of the transcription factor in their proximal regulatory regions (FDR < 0.01).
2.4. Survival analyses
Kaplan–Meier survival curves were plotted using the R package survminer, in which high and low patient groups were divided by the median. Log rank tests were used to compare the survival of patient groups. Hazard ratios were estimated by univariate Cox proportional hazard regression and LASSO Cox regression. Wald tests were performed to calculate the significance of the univariate Cox regression models (p-value < 0.05). The log rank tests and the univariable Cox regression were accomplished by survival [27]. The LASSO Cox regression was performed by glmnet [28].
The risk score of the selected transcription factors was calculated as follows:
where Li is the coefficient of the ith transcription factor calculated by the LASSO Cox regression, and Xi is the Z-score transformed activity of the ith transcription factor.
2.5. Statistical analyses
Differences between two groups was analyzed by two tailed t-tests, except for comparisons of transcription factor activity between prostate tumors and normal prostates. To compare more than two groups, one-way ANOVA was first performed. If the ANOVA test was significant (p-value < 0.05), then a Tukey's HSD test was performed for multiple pairwise comparisons.
2.6. Functional analyses
Gene Set Enrichment Analyses (GSEA) and over-representation analyses were performed using clusterProfiler [29].
2.7. Data visualization and database construction
The data visualization tools used in this study included Cytoscape [30] and R packages such as ggplot2 [31], ggsignif, ComplexHeatmap [32], and clusterProfiler [29].
The online LNCTRN database was developed using the R package shiny, which collected transcriptional circuits governed by transcription factors and lncRNAs.
2.8. RNA immunoprecipitation quantitative PCR (RIP-qPCR)
Native RIP was performed as previously described [33] in the LNCaP prostate cancer cells (American Type Culture Collection CRL-1740) using 5 μg of antibodies against MYC (ab32072), AR (ab74272), YBX1 (20339-1-AP), or IgG, except that immunoprecipitated and input RNAs were purified by the TRIzol reagent (Invitrogen) following the manufacturer's protocol. RIP with anti-IgG served as a negative control. Reverse transcription was conducted using NovoScript 1st Strand cDNA Synthesis SuperMix (Novoprotein), and quantitative PCR was performed in triplicate using AceQ qPCR SYBR Green Master Mix (Vazyme Biotech). Primer sequences were listed in supplementary Table S5.
2.9. Cell transfection
Synthesized siRNAs (GenePharma) were transfected into the LNCaP cells using HilyMax (Dojindo). Sequences of siRNAs were listed in supplementary Table S6. Cells were harvested six hours following transfection, and then total RNAs were extracted by TRIzol. Quantitative reverse PCR was performed as described in RIP-qPCR using the primers shown in supplementary Table S5. Actin was used as the reference gene.
2.10. Role of funding source
Financial support was provided by National Natural Science Foundation of China and Fudan University. None of these sponsors had any role in the study design, the collection, analysis, and interpretation of data, the writing of the report, or the decision of paper submission.
3. Results
3.1. Transcription factor activity in prostate tumors and normal prostates
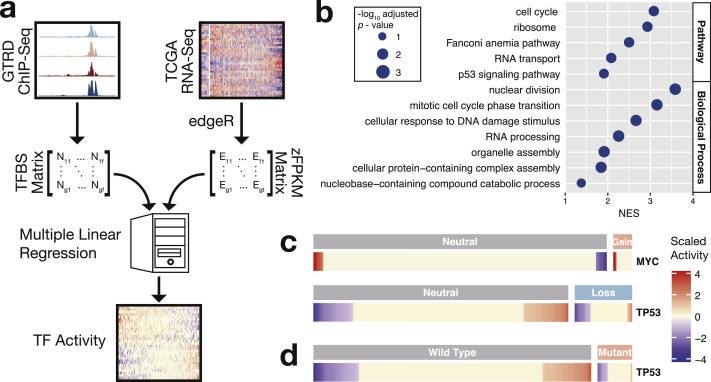
An increasing number of studies are focusing on the roles of transcription factors in driving and promoting the initiation and progression of prostate cancer [1,2]. Their activities rely not only on expression levels but also on protein modifications, such as phosphorylation, which may alter transcription factor stability, structure, localization, and interactome [34]. Furthermore, their transcriptional effects usually necessitate cooperation of multiple factors and are dependent on cellular contexts [34,35]. In the present study, a multivariate regression model [36] was adopted to estimate transcription factor activity. In brief, a multiple linear regression analysis was performed in each sample. In the analyses, the transcription factor occupancies were regarded as the predictor variables and the gene expression levels (standardized fragments per kilobase per million mapped fragments [zFPKM]) as the outcome variables (Fig. 1a). Given that the present study focused on transcription factors potentially involved in prostate carcinogenesis, the regression analyses were restricted to genes showing differential expression between primary tumors and normal tissues. Regression coefficients are estimates of transcription factor activity. To ascertain whether our regression model could infer transcription factor activity, we first investigated the two well-studied tumor proteins MYC and TP53. MYC acts as a master regulator of cell growth by orchestrating the expression of genes involved in cell cycle, DNA replication, ribosome biosynthesis, and other metabolisms [10]. The characteristic genes appeared at the top of the gene list ranked by expression correlations with MYC activity (Fig. 1b and Supplementary Fig. S1). In addition, MYC activity increased with gene copy gains (p-value = 4E-6, Fig. 1c). By contrast, the activity of the other transcription factor, TP53, which is also known as a p53 tumor suppressor, decreased with gene copy losses (p-value = 5E-5, Fig. 1c). TP53 is one of the most commonly mutated proteins in human cancers, including prostate cancer [7,37], even though somatic mutations are far less frequent in prostate cancer than in other solid tumors [7]. Notably, the mutations led to reduced TP53 activity (p-value = 3E-3, Fig. 1d), albeit with no change in expression levels (p-value > 0.05), a result consistent with that of a previous report [38]. Moreover, not all transcription factor activities were influenced by genomic copy number alterations (data not shown). Copy number variations may change gene expression levels [1]. However, expression level is not the only determinant of transcription factor activity [34,35]. This fact may explain the discrepancy between genetic copy variations and the activity of the transcription factors.
Fig. 1.
Calculation of transcription factor activity in normal prostates and primary prostate cancers. (a) Computational workflow of transcription factor activity. In brief, multiple linear regression was applied to the zFPKM values of a gene set and binding site numbers of transcription factors in the proximal regulatory regions of the same gene set. The selected set of genes were those differentially expressed between tumors and normal tissues. Regression coefficients estimated transcription factor activity. (b) Biological roles of the genes showing expression correlation with MYC activity (BH adjusted p-value < 0.05), which were in accordance with the reported functions of MYC. (c) Genomic copy number variations and (d) mutations affected TP53 and MYC activity. Mutations included deleterious mutations, missense, and in-frame INDELs. zFPKM, standardized fragments per kilobase per million mapped fragments.
The activity of 67 and 63 transcription factors increased and decreased in prostate cancer, respectively (Figs. 2a–2e and Supplementary Table S1). The reduced activity of the TP53 tumor suppressor (Fig. 2b) and the elevated activity of the MYC proto-oncogene (Fig. 2c) in primary tumors were expected. The enhanced AR activity observed in the tumors (Fig. 2a) agreed with the previous findings that the AR signaling pathway is a central axis in promoting the growth and survival of both normal prostate cells and prostate cancer cells [1,2,4,5]. A prominent example of the role of AR in prostate cancer initiation is the androgen-dependent activation of ERG. Recurrent fusion of the androgen-dependent TMPRSS2 promoter and ERG and thereby ERG overexpression drive prostate cancer cell invasion [8]. Indeed, ERG activity increased in the primary tumors (Fig. 2d). Furthermore, disease ontology analysis of the above 130 transcription factors showing differential activity between tumors and normal tissues revealed that almost all of the enriched terms were regarding cancer (Fig. 2f and Supplementary Fig. S2), and prostate cancer ranked at the top. In addition, the pathway of transcriptional misregulation in cancer was overrepresented (Fig. 2g).
Fig. 2.
Differential transcription factor activity between normal prostates and primary prostate cancers. (a–e) Activity profiles of transcription factors activated/repressed in primary prostate cancers compared with those in normal prostates. Activity changes are displayed in the rightmost column. (f) Disease ontologies (top three) and (g) KEGG pathways enriched in the altered transcription factors (BH adjusted p-value < 0.05). TF, transcription factor; FC, fold change.
Overall, the multiple linear regression model approximated transcription factor activity on the basis of the activity profiles of the key transcription factors participating in the prostate cancer pathogenesis, such as MYC, TP53, AR, and ERG, which was confirmed by comprehensive in silico functional analyses.
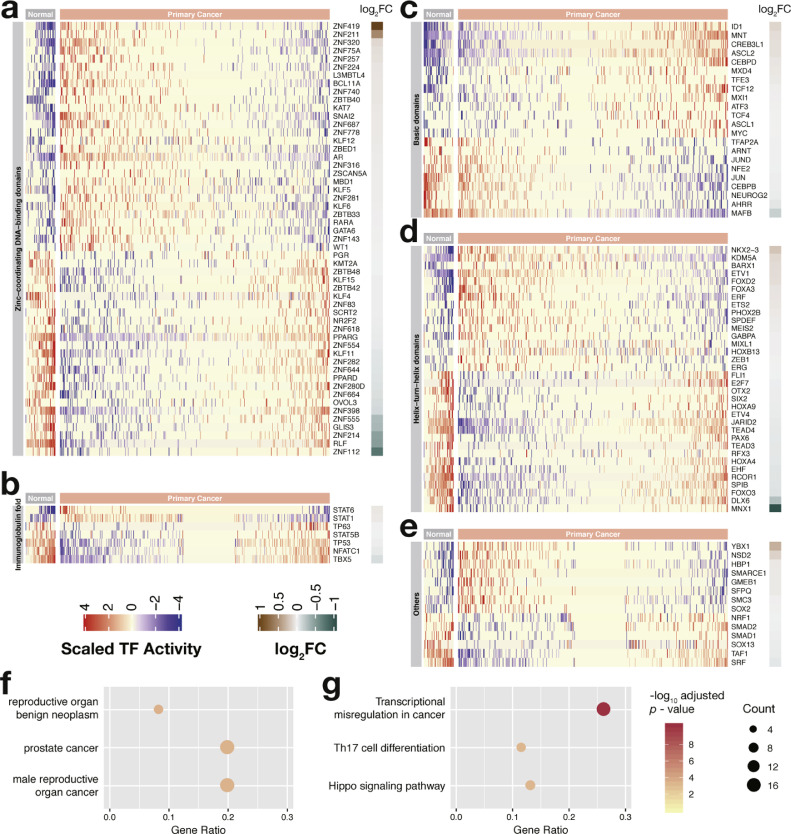
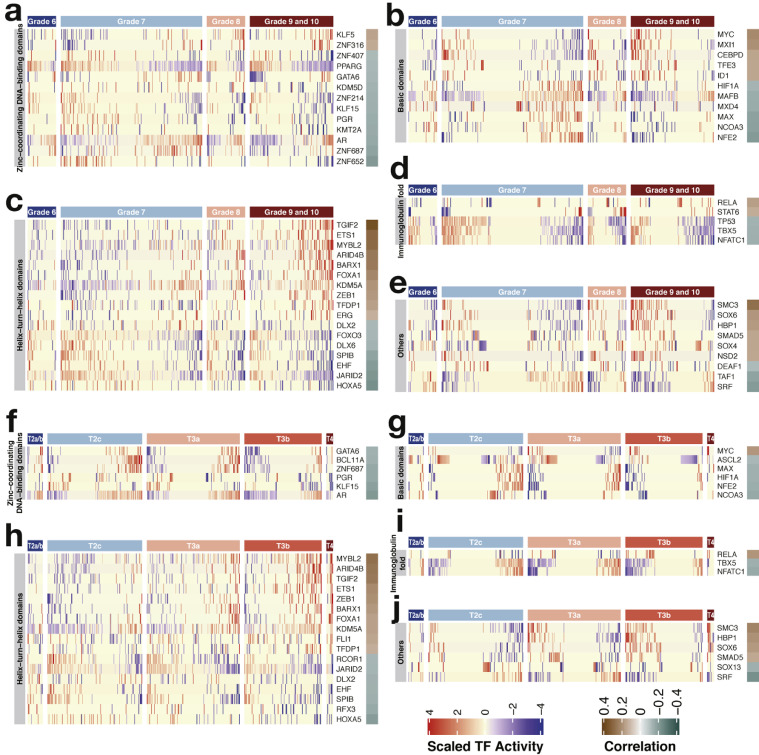
3.2. Transcription factors associated with the clinicopathological characteristics of prostate cancer
The clinical value of the transcription factors was then evaluated by calculating the Pearson correlation between their activity and the clinicopathological factors of prostate cancer. The Gleason score is a grading system of prostate cancer that ranges from 6 to 10; in this system, cancers with a higher score are more aggressive [39]. A total of 26 and 30 transcription factors were positively and negatively correlated with the histologic grading, respectively (Figs. 3a–3e and Supplementary Table S2). The TNM staging system is also commonly used to determine the malignancy of prostate cancer [39]. Herein, we focused on clinical T staging which describes the size and extent of primary tumors. Prostate cancers are mainly classified into T1, T2, T3, and T4. A larger number after T indicates a larger tumor and a wider spread to nearby tissues. Positive and negative correlations with the T stage were found in 16 and 22 transcription factors, respectively (Figs. 3f–3j and Supplementary Table S3). MYC, TGIF2, ETS1, MYBL2, ARID4B, BARX1, FOXA1, KDM5A, ZEB1, TFDP1, RELA, SMC3, SOX6, HBP1, and SMAD5 were positively correlated with both histologic grading and clinical staging. By contrast, AR, GATA6, KLF15, PGR, ZNF687, HIF1A, MAX, NCOA3, NFE2, DLX2, SPIB, EHF, JARID2, HOXA5, TBX5, NFATC1, and SRF were negatively correlated with both clinicopathological parameters. These transcription factors might be indicative of cancer malignancy. Intriguingly, AR activity declined in advanced tumors. AR is generally thought to promote the growth and survival of prostate cancer cells [1,2,5]. However, a recent study has demonstrated that overactive AR induces DNA double-strand breaks and cell cycle arrest [40], which indicates the complex roles of AR in prostate cancer.
Fig. 3.
Activity profiles of transcription factors correlated with the clinicopathological characteristics (FDR < 0.05). (a–e) Activity profiles of transcription factors correlated with histologic grading. (f–j) Activity profiles of transcription factors correlated with clinical staging. Samples are arranged from left to right in ascending order of aggressiveness. Pearson correlation coefficients between transcription factor activity and grading/staging are displayed in the rightmost column. TF, transcription factor.
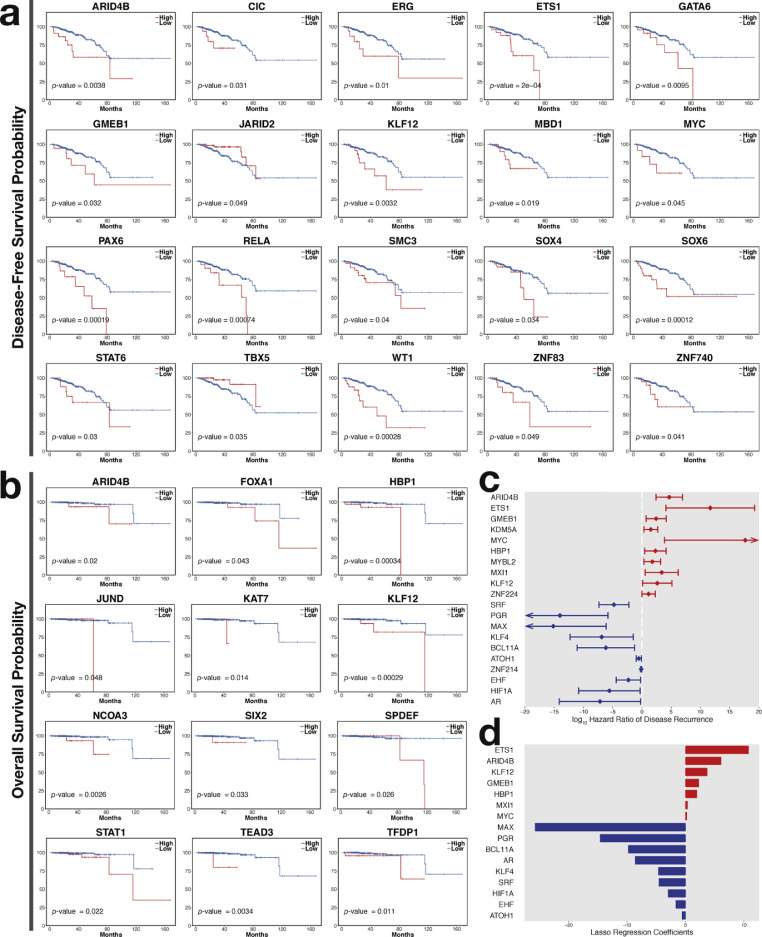
3.3. Prognostic transcription factors for prostate cancer
Various risk stratification systems have been developed to assist the decision-making process in prostate cancer treatment [39]. A scheme that incorporates traditional clinicopathological parameters performs well, but uncertainty about progression risk still persists. Several newly developed molecular tools have been demonstrated to improve the accuracy of prognosis. In this study, Kaplan–Meier survival analyses were performed on transcription factor activity to discover novel biomarkers. A total of 20 transcription factors, namely, ARID4B, CIC, ERG, ETS1, GATA6, GMEB1, JARID2, KLF12, MBD1, MYC, PAX6, RELA, SMC3, SOX4, SOX6, STAT6, TBX5, WT1, ZNF740, and ZNF83, were strongly associated with prostate cancer relapse (Fig. 4a). Less significant relevance was found in GMEB2, HIC1, HOXA5, KLF6, MAX, MNT, PGR, RUNX2, SPI1, SRF, TFDP1, and ZNF664 (Supplementary Fig. S3a). Among these transcription factors, high levels of JARID2, TBX5, HOXA5, MAX, PGR, RUNX2, SRF, and ZNF664 favored a good prognosis. The potential prognostic factors for overall survival included ARID4B, FOXA1, HBP1, JUND, KAT7, KLF12, NCOA3, SIX2, SPDEF, STAT1, TEAD3, and TFDP1 (Fig. 4b). The less reliable ones included DMAP1, MYC, NRF1, and WT1 (Supplementary Fig. S3b), all of which implied a high risk of death. Moreover, univariate Cox regression analyses for cancer recurrence revealed that ARID4B, ETS1, GMEB1, KDM5A, MYC, HBP1, MYBL2, MXI1, KLF12, and ZNF224 were poor prognostic factors, whereas SRF, PGR, MAX, KLF4, BCL11A, ATOH1, ZNF214, EHF, HIF1A, and AR were good ones (Fig. 4c and Supplementary Fig. S4b). In terms of risk of death, HBP1, KLF12, WT1, ZNF143, TGIF2, TFDP1, and MYC implied poor outcomes, whereas OVOL3, SPIB, HOXA5, NFATC1, MAX, SRF, and ATF3 suggested beneficial effects (Supplementary Fig. S4a).
Fig. 4.
Potential transcription factors essential for prostate cancer progression. (a) Disease-free and (b) overall survival associated transcription factors (p-value < 0.05). (c) Hazard ratios and 95% confidence intervals of hazard ratios of transcription factors in relation to disease-free survival (p-value < 0.05). The x-axis is truncated at −20 and 20. (d) LASSO Cox regression coefficients of transcription factors in relation to disease-free survival. In (c) and (d), a red transcription factor indicates a bad prognosis, whereas a blue transcription factor denotes a protective effect.
A LASSO Cox regression model was applied to remove the transcription factors with minimal effects on patient survival, build a prognostic signature, and predict clinical outcomes better. Sixteen transcription factors, namely, ETS1, ARID4B, KLF12, GMEB1, HBP1, MXI1, MYC, MAX, PGR, BCL11A, AR, KLF4, SRF, HIF1A, EHF, and ATOH1, were selected to construct the prognostic signature for prostate cancer relapse (Fig. 4d). However, no transcription factor was identified to independently contribute to the overall survival. The mean follow-up time after radical prostatectomy is less than 2 years, which limits the survival analysis because of the long natural history of prostate cancer [7]. We focused on these prognostic transcription factors hereafter. Among them, MYC, MAX, and MXI1 share an identical DNA binding motif (position weight matrix) [41]. Thus, our analysis might not have been able to differentiate them or even identify other paralogous transcription factors. However, no correlation was observed among the activities of MYC, MAX, and MXI1 in prostate cancer, except for a moderate correlation between MYC and MXI1 (Pearson Correlation = 0.17 and 0.01 < false discovery rate < 0.05). In fact, although paralogous transcription factors often share similar/identical DNA binding motifs, they have intrinsic differences in DNA binding preferences that contribute to their distinct in vivo binding [42].
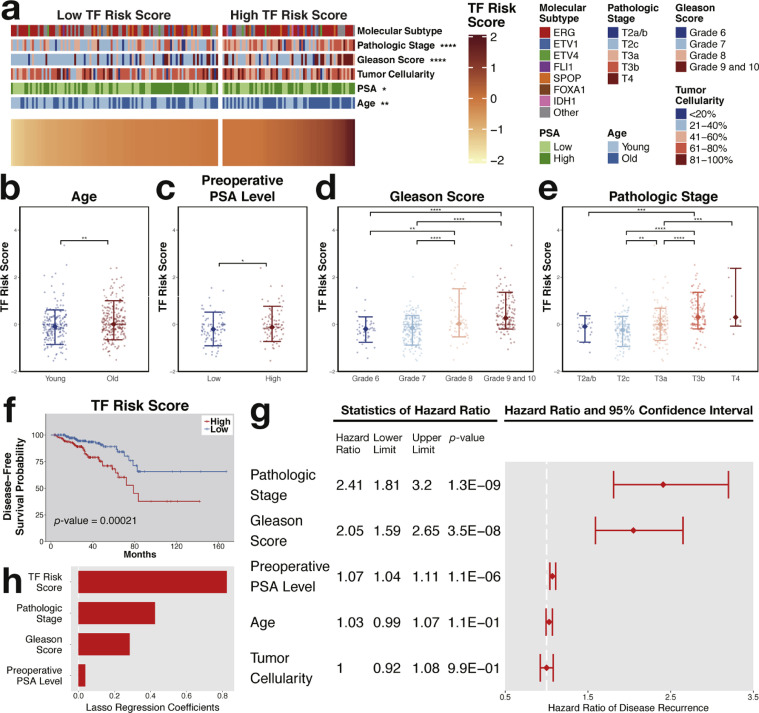
Risk scores were calculated on the basis of the 16-gene prognostic signature. The Gleason score, clinical stage, age, tumor cellularity, and PSA level are all associated with the risk of prostate cancer [39]. Higher risk scores were observed in the patients of older age, higher preoperative PSA levels, higher Gleason scores, or higher T stages who are actually at a higher risk for prostate cancer (Figs. 5a–5e). However, no difference was detected in tumor cellularity in terms of the risk score (Supplementary Fig. S5a). Overall, the risk score agreed well with the clinicopathological factors. The TCGA research group defines seven distinct molecular subtypes in prostate cancer according to genomic profiling [7]. However, there seemed to be no relationship between the subtype and the risk score (Supplementary Fig. S5b). Further Kaplan–Meier survival analysis suggested that the risk score of the 16-transcription factor signature could be a prognostic factor (Fig. 5f). Additional univariate and multivariate Cox regression analyses confirmed the prognostic value of the 16-transcription factor signature and could independently predict patient survival (Figs. 5g and 5h).
Fig. 5.
Prognostic value of the risk transcription factors. (a) Clinicopathological differences between groups of low and high risk scores. Risk scores increased in the patients older in age (b) and higher in preoperative PSA levels (c), histologic grade (d), and T stage (e). (f) Worse disease-free survival was observed in the group with a higher risk score (p-value < 0.01). (g) Hazard ratios and 95% confidence intervals of hazard ratios of clinicopathological characteristics. (h) LASSO Cox regression coefficients of risk factors in relation to disease-free survival. TF, transcription factor. * denotes p-value < .05; ** indicates p-value < .01; *** signifies p-value < .001; **** represents p-value < .0001.
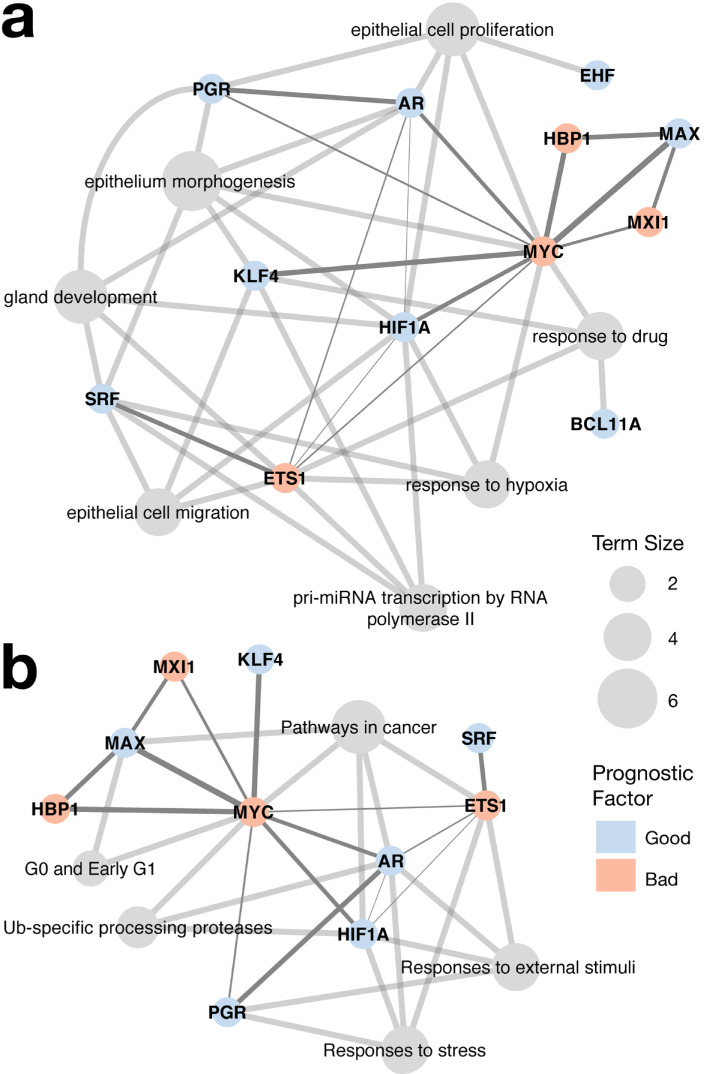
More than half of the prognostic signature constituent transcription factors have been implicated in prostate cancer (Supplementary Table S4). Except for those of BCL11A and HIF1A, their effects on survival were consistent with their reported roles in cancer probably because of the complexity of gene interaction and cancer heterogeneity. For example, MAX interacts with not only pro- but also anti-oncogenic factors [41]. As another example, ATOH1 prevents gastric tumor formation [43] but promotes medulloblastoma growth [44]. These signature transcription factors participated in gland development, epithelium morphogenesis, epithelial cell proliferation and migration, response to hypoxia and drug, and pri-miRNA transcription by RNA polymerase II (Fig. 6a), aside from cancer pathways, G0 and early G1, Ub-specific processing proteases, and response to stress and external stimuli (Fig. 6b).
Fig. 6.
Network regulated by the prognostic signature constituent transcription factors. (a) Biological processes and (b) KEGG pathways governed by the signature transcription factors. The edges linking transcription factors are marked in dark grey and their width is scaled according to the combined interaction scores between transcription factors calculated by the STRING database [55].
3.4. Cis- and trans-regulation by long noncoding RNAs in prostate cancer
Another type of transcriptional regulator is long noncoding RNA which can act either in cis (adjacent to its transcription start site) or in trans (distal to it transcription start site) [13,14]. The involvement of lncRNAs as transcriptional regulators in carcinogenesis has been demonstrated in certain primary cancers, including prostate cancer [20,21,45]. Herein, we constructed a transcriptional network to decipher the lncRNA regulation of dysregulated genes in prostate cancer.
A total of 42 cis-regulations by lncRNAs that involved 41 lncRNAs were identified on the basis of expression correlation and distance between gene loci (Supplementary Fig. S6a).
Trans-regulation by lncRNAs might be achieved by its direct interaction with genomic regulatory elements and/or by binding to other transcriptional regulators such as transcription factors [13,14]. For the first type of regulation, we focused the analysis on lncRNAs that might form lncRNA–DNA triplexes in transcription regulatory regions (Supplementary Fig. S6b). Accumulating evidence provides more insights into the RNA–DNA triplex formation as an important mechanism of transcriptional regulation by lncRNAs [46]. In sum, 9474 trans-regulations through lncRNA–DNA triplexes that involved 531 lncRNAs were identified.
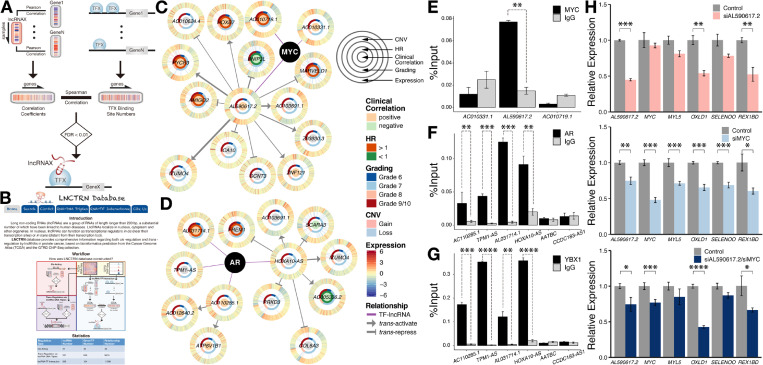
LncRNAs can bind to transcription factors to alter their activity, stability, localization, and interacting partners [13,14]. We developed a pipeline to recognize interactions among lncRNAs and transcription factors (Fig. 7a) on the basis of the hypothesis that lncRNAs can regulate gene transcription by interacting with transcription factors. Therefore, an lncRNA and a transcription factor should share common target genes if they can interact with each other. In our analysis, the common target genes were confined to the transcription factor targets also showing differential expression in prostate cancer. The occupancies of the transcription factors in the proximal regulatory regions of the common target genes were obtained to approximate their effects on the target expression. Pearson correlations with the common target genes in the expression level were also computed for the lncRNAs to estimate their regulation of the targets. Finally, Spearman's rank correlation was calculated between lncRNA regulation and transcription factor effects to explore their interactions. In total, 11398 interactions were found that included 839 lncRNAs and 124 transcription factors.
Fig. 7.
Long noncoding RNAs interacting with the prognostic signature constituent transcription factors. (a) Prediction pipeline of transcription factor interaction with lncRNAs by measuring the correlation between lncRNA regulation of gene expression and transcription factor occupancies in proximal regulatory regions among differentially expressed transcription factor targets. (b) The LNCTRN database provides comprehensive information of both cis- and trans-regulations by lncRNAs in prostate cancer. (c and d) A glance at the transcriptional network governed by the MYC– and AR–lncRNA interactions. Each gene is depicted as a multi-ring circle listing expression correlation (Pearson) with histologic grading, hazard ratio of disease recurrence, tumor grades, copy number variations, and zFPKM values from the inner to the outer layer. The outermost three datasets are plotted in the way that each “spoke” represents a single sample. Samples are arranged in the same order for all genes. (e) MYC-interacting lncRNAs displayed in (c) were validated by RIP-qPCR in LNCaP prostate cancer cells. (f) AR-interacting lncRNAs exhibited in (d) were validated by RIP-qPCR in LNCaP cells, in addition to their interactions with YBX1 (g). (h) Effects of AL590617.2 (siAL590617.2), MYC (siMYC), and AL590617.2/MYC (siAL590617.2/siMYC) knockdown on AL590617.2 and MYC candidate targets in LNCaP cells. * denotes p-value < .05; ** indicates p-value < .01; *** signifies p-value < .001; **** represents p-value < .0001.
3.5. LNCTRN database for lncRNAs involved in transcriptional circuits
The aforementioned regulatory circuits in which lncRNAs participate were recorded in our database named LNCTRN (https://navy.shinyapps.io/lnctrn) (Fig. 7b and Supplementary Figs. S7a–S7c). All three types of relationships are represented in the form of a chart in which the first two columns list Ensembl IDs and official symbols of lncRNAs, the next two columns register Ensembl IDs and official symbols of lncRNA-regulated genes or interacting transcription factors, and the last two columns itemize the estimates of relationship strength by correlation analyses. In addition, distances between the transcription start sites of lncRNAs and their target genes are shown for the cis-regulation relationships. A click on an Ensembl ID will trigger the redirection to its gene record. Entering a string in the search box above a table will enable partial matching and return rows containing the string. Furthermore, a simple gene search is provided that supports a single query of either an Ensembl ID or an official symbol (Supplementary Fig. S7d).
In each gene record, the following pieces of information are listed from top to bottom: basic gene information, subcellular localization, correlations with histologic grading and clinical staging, linkage with survival, and transcriptional regulations involved (Supplementary Fig. S7e). In the section of basic gene information, the official symbol, Ensembl ID, full name, and chromosomal location are displayed, in addition to a boxplot of expression levels in prostate tumors and normal prostates. An external link to Ensembl is added to the Ensembl ID for further gene information. In the section of subcellular localization, the database provides subcompartment enrichment on the basis of cytoplasmic and nuclear RNA-Seq and other types of experimental evidence obtained from public resources [47,48]. In the sections of correlations with histologic grading and clinical staging, expression differences between grades, between T stages, and between N stages are displayed. Comparison between M0 and M1 was skipped because of the limited number of M1 samples (only three). In the section of survival analyses, association with death and tumor recurrence are shown. In the sections of transcriptional regulations, all types of relationships are listed in tables, and links to respective gene pages are also provided. With lncRNA AP006284.1 as an illustrative example, the following pieces of information can be obtained from LNCTRN: (1) Ensembl ID is ENSG00000254815 and full name is LMNTD2 antisense RNA 1, which is located on the positive strand of chromosome 11 from position 557,595 to 560,107; (2) mean FPKM values are 1.7 and 0.6 in prostate cancer and normal prostate, respectively; (3) its expression is enriched in nucleus; (4) expression levels increase in tumors of high grade (8 to 10) and advanced stage (T3b and N1); (5) higher expression level is significantly linked to a shorter relapse period and moderately associated with shorter overall survival time; (6) it probably regulates two genes in cis; (7) it may regulate one gene in trans by forming an RNA–DNA triplex near the transcription start site of the target gene; and (8) it may function as a transcriptional regulator by interacting with one or more of 27 transcription factors.
3.6. Transcriptional network governed by the signature transcription factor–lncRNA interactions
We focused our study on the prognostic transcription factors and their lncRNA partners. Among the 16 transcription factors, MYC and AR were hub genes showing high connectivity (Fig. 6). We then narrowed down our focus to MYC and AR. In total, 201 and 147 lncRNAs were identified to potentially interact with MYC (Supplementary Fig. S8a) and AR (Supplementary Fig. S8b), respectively. At least three MYC and AR interacting lncRNA candidates were randomly selected for experimental validation (Figs. 7c and 7d).
One out of the three MYC candidates were confirmed to physically interact with MYC (Fig. 7e). AL590617.2 expression increased with the advance of tumor grade (Fig. 7c). It might probably either form a complex with MYC to transcriptionally regulate MYC targets or trans-regulate genes such as MARVELD1, HOXB7, PYCR3, AMIGO2, BNIP3L, and ZNF121 by forming an lncRNA–DNA triplex in the regulatory regions of the target genes followed by MYC recruitment (Fig. 7c).
Four randomly selected lncRNA partners of AR, namely, AC110285.1, TPM1-AS, AL031714.1, and HOXA10-AS, were confirmed (Fig. 7f). AL031714.1 and AC110285.1 exhibited elevated expression in high-grade tumors, and patients with high AL031714.1 levels were prone to bad prognoses (Fig. 7d). YBX1 can interact with AR and function as an AR activator in LNCaP prostate cancer cells [49]. Co-immunoprecipitation of AC110285.1, TPM1-AS, AL031714.1, and HOXA10-AS with YBX1 (Fig. 7g) confirmed the interactions between AR and the lncRNAs HOXA10-AS, AC110285.1, TPM1-AS, and AL031714.1 which might cooperate with AR to regulate the transcription of the AR targets. The first two lncRNAs might be able to trans-regulate genes, such as AC005336.2, SCARA3, FREM1, ATP6V1B1, and AC012640.2, by forming lncRNA–DNA hybrids in the regulatory regions of the target genes and may further recruit AR (Fig. 7d).
Transcription regulation by MYC–AL590617.2 was further explored. We performed RNA interference targeting AL590617.2 and MYC and examined expression changes in the top four AL590617.2-correlated MYC target genes (Supplementary Excel S1). AL590617.2 and MYC knockdown substantially reduced the expression of two targets, namely, OXLD1 and REX1BD (Fig. 7h). In addition, MYC knockdown decreased AL590617.2 expression levels. Thus, MYC and AL590617.2 may form a feed-forward regulation loop that is a widespread strategy of transcriptional control [50].
4. Discussion
Over the past decades, earlier detection and treatment have accounted for over 50% reduction in deaths due to prostate cancer [39]. Beyond the traditional diagnosis scheme that considers Gleason grade, stage, and PSA, the use of molecular biomarker assays has greatly enhanced the sensitivity and specificity of prostate cancer detection and is believed to provide more accurate guidelines for optimal treatment choice [39]. By combining transcriptomic, epigenetic, and clinicopathological data, we constructed a prognostic signature consisting of 16 transcription factors, namely, ETS1, ARID4B, KLF12, GMEB1, HBP1, MXI1, MYC, MAX, PGR, BCL11A, AR, KLF4, SRF, HIF1A, EHF, and ATOH1. The score of the signature could reflect prostate cancer malignancy and independently predict recurrence risk. Thus, the prognostic signature may serve as a marker for prostate cancer progression. Out of the 16 signature transcription factors, AR and MYC are known as prostate cancer drivers [1,2,5,11,12]. In addition, ETS1, KLF12, MAX, BCL11A, SRF, and EHF are regarded as pivotal prostate cancer regulators as reported by Dhingra, et al. who integrated single nucleotide variation, structural variation, DNA methylation, and DNase I hypersensitive sites and discovered 153 transcription factors potentially playing a key role in the transcriptional network of prostate cancer [51]. Furthermore, the signature transcription factors were implicated in gland development and epithelium morphogenesis. Dedifferentiation of functional mature cells into progenitor-like cells is generally regarded as an essential early step in cancer initiation [52]. Development and carcinogenesis usually share a set of machinery, such as transcription factors many of which participate in not only cellular identity specification but also oncogenic dedifferentiation [52]. The involvement of signature transcription factors in gland development and epithelium morphogenesis implies the importance of these factors in prostate cancer. Human oncogenes are considerably more numerous than oncogenic transcription factors, making transcription factors ideal for therapeutic targeting [2]. Except for AR, MYC, and MAX that have had approved targeting drugs [1,2,5,6], the prognostic signature constituent transcription factors, especially the oncogenic ones, can be novel candidates for drug development in prostate cancer.
Emerging evidence increasingly highlights the influence of lncRNA interplay with transcription factors on fine-tuning transcriptional programs [13,14]. In the current study, 11398 interactions were discovered between 839 lncRNAs and 124 transcription factors in primary prostate cancer by integrating the transcriptomic data of primary prostate cancer and transcription factor-binding profiles. To the best of our knowledge, the present study is the first to identify the interaction between transcription factors and lncRNAs in a large scale. Interactions involving MYC, AR, and the AR activator YBX1 were randomly checked, and many of them were experimentally confirmed. Thus, our pipeline can be applied to other tissues or cancer types to recognize interactions among lncRNAs and transcription factors. However, the interactions identified in our study are still “correlations” in essence. Under the circumstance that a transcription factor exerts similar effects on regulating the expression levels of an lncRNA and other targets, our algorithm may consider an interaction that exists between the transcription factor and the lncRNA even though there is no interaction in fact. Thus, given the complexity of transcriptional regulation, the interactions should be contemplated with caution.
Transcription factor-bound lncRNAs may enhance or attenuate the transcription of target genes by altering transcription factor activity and stability [13,14]. Moreover, lncRNAs may directly target gene regulatory regions, subsequently recruiting their transcription factor partners [13,14]. Exploring the transcriptional regulation by lncRNAs aided in not only illustrating the full transcriptional network but also identifying novel oncogenes and tumor suppressors. The interacting lncRNA partners of the key transcription factors to cancer can be reasonably implicated in cancer development. Indeed, among the AR-interacting lncRNAs experimentally proved in this study, AL031714.1 and AC110285.1 expression correlated with tumor malignancy, and high AL031714.1 levels contributed to a bad prognosis. Moreover, HOXA10-AS was recently demonstrated to promote cell proliferation of lung adenocarcinoma [53] and KMT2A-rearranged leukemia [54], although its expression was not found to correlate with either tumor grade or survival in prostate cancer. Thus, the interacting lncRNA partners of the prognostic signature constituent transcription factors may represent potential biomarkers and/or therapeutic targets in prostate cancer.
In conclusion, we identified 16 transcription factors whose combined activity can be an independent prognostic factor for prostate cancer, in addition to their interacting lncRNA partners. Further investigation on these transcriptional regulators may provide a path toward the development of new therapeutic targets. Moreover, the regulatory network governed by the transcription factors and their interacting lncRNA partners were illustrated and stored in the LNCTRN database. The computational framework proposed in our study can be applied to discover critical transcriptional regulators in other cancer types.
Data sharing
Copy number variation, genomic mutation, gene expression, and clinicopathological profiles of primary prostate tumours and/or normal prostates were downloaded from the GDC data portal of the Cancer Genome Atlas (TCGA, https://portal.gdc.cancer.gov). Molecular subtype, preoperative PSA level, and tumour cellularity of primary prostate tumours were obtained through TCGAbiolinks [24]. Transcription factor-binding profiles were downloaded from the GTRD ChIP-Seq database [22].
Our LNCTRN database records comprehensive information of regulatory network governed by transcription factors and their interacting long noncoding RNA partners and can be explored at (https://navy.shinyapps.io/lnctrn).
Declaration of Competing Interest
The authors declare that they have no competing interests.
Acknowledgments
Acknowledgements
This study was supported by National Natural Science Foundation of China (31571330, 81773024, and 31701282) and Open Research Fund of the State Key Laboratory of Genetic Engineering, Fudan University (SKLGE-1901).
Author contributions
YL and MJ conceived the study; YHC and MJ collected the data; MJ, YHC, SHG, CJW, and YH analyzed and interpreted the data; DW and YLL performed the experiments; MJ wrote the manuscript. All authors reviewed and approved the final manuscript.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ebiom.2020.103150.
Contributor Information
Mei Jiang, Email: 042023070@fudan.edu.cn.
Yao Li, Email: yaoli@fudan.edu.cn.
Appendix. Supplementary materials
References
- 1.Baumgart SJ, Nevedomskaya E, Haendler B. Dysregulated transcriptional control in prostate cancer. Int J Mol Sci. 2019;20:2883. doi: 10.3390/ijms20122883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bushweller JH. Targeting transcription factors in cancer - from undruggable to reality. Nat Rev Cancer. 2019;19:611–624. doi: 10.1038/s41568-019-0196-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hazelett DJ, Rhie SK, Gaddis M. Comprehensive functional annotation of 77 prostate cancer risk loci. PLoS Genet. 2014;10 doi: 10.1371/journal.pgen.1004102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wilson JD. The critical role of androgens in prostate development. Endocrinol Metab Clin North Am. 2011;40:577–590. doi: 10.1016/j.ecl.2011.05.003. ix. [DOI] [PubMed] [Google Scholar]
- 5.Tan MH, Li J, Xu HE, Melcher K, Yong EL. Androgen receptor: structure, role in prostate cancer and drug discovery. Acta Pharmacol Sin. 2015;36:3–23. doi: 10.1038/aps.2014.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wade CA, Kyprianou N. Profiling prostate cancer therapeutic resistance. Int J Mol Sci. 2018;19:904. doi: 10.3390/ijms19030904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cancer Genome Atlas Research N The molecular taxonomy of primary prostate cancer. Cell. 2015;163:1011–1025. doi: 10.1016/j.cell.2015.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nicholas TR, Strittmatter BG, Hollenhorst PC. Oncogenic ETS factors in prostate cancer. Adv Exp Med Biol. 2019;1210:409–436. doi: 10.1007/978-3-030-32656-2_18. [DOI] [PubMed] [Google Scholar]
- 9.Rickman DS, Soong TD, Moss B. Oncogene-mediated alterations in chromatin conformation. Proc Natl Acad Sci U S A. 2012;109:9083–9088. doi: 10.1073/pnas.1112570109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dang CV. MYC, metabolism, cell growth, and tumorigenesis. Cold Spring Harb Perspect Med. 2013;3 doi: 10.1101/cshperspect.a014217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rebello RJ, Pearson RB, Hannan RD, Furic L. Therapeutic approaches targeting MYC-driven prostate cancer. Genes (Basel) 2017;8:71. doi: 10.3390/genes8020071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Labbe DP, Brown M. Transcriptional regulation in prostate cancer. Cold Spring Harb Perspect Med. 2018;8 doi: 10.1101/cshperspect.a030437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun Q, Hao Q, Prasanth KV. Nuclear long noncoding RNAs: key regulators of gene expression. Trends Genet. 2018;34:142–157. doi: 10.1016/j.tig.2017.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yao RW, Wang Y, Chen LL. Cellular functions of long noncoding RNAs. Nat Cell Biol. 2019;21:542–551. doi: 10.1038/s41556-019-0311-8. [DOI] [PubMed] [Google Scholar]
- 15.Chakravarty D, Sboner A, Nair SS. The oestrogen receptor alpha-regulated lncRNA NEAT1 is a critical modulator of prostate cancer. Nat Commun. 2014;5:5383. doi: 10.1038/ncomms6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yap KL, Li S, Munoz-Cabello AM. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol Cell. 2010;38:662–674. doi: 10.1016/j.molcel.2010.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang L, Lin C, Jin C. lncRNA-dependent mechanisms of androgen-receptor-regulated gene activation programs. Nature. 2013;500:598–602. doi: 10.1038/nature12451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Prensner JR, Sahu A, Iyer MK. The IncRNAs PCGEM1 and PRNCR1 are not implicated in castration resistant prostate cancer. Oncotarget. 2014;5:1434–1438. doi: 10.18632/oncotarget.1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang A, Zhao JC, Kim J. LncRNA HOTAIR enhances the androgen-receptor-mediated transcriptional program and drives castration-resistant prostate cancer. Cell Rep. 2015;13:209–221. doi: 10.1016/j.celrep.2015.08.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arun G, Diermeier SD, Spector DL. Therapeutic targeting of long non-coding RNAs in cancer. Trends Mol Med. 2018;24:257–277. doi: 10.1016/j.molmed.2018.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Oliveira JC, Oliveira LC, Mathias C. Long non-coding RNAs in cancer: another layer of complexity. J Gene Med. 2019;21:e3065. doi: 10.1002/jgm.3065. [DOI] [PubMed] [Google Scholar]
- 22.Yevshin I, Sharipov R, Valeev T, Kel A, Kolpakov F. GTRD: a database of transcription factor binding sites identified by ChIP-seq experiments. Nucleic Acids Res. 2017;45:D61–DD7. doi: 10.1093/nar/gkw951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mounir M, Lucchetta M, Silva TC. New functionalities in the TCGAbiolinks package for the study and integration of cancer data from GDC and GTEx. PLoS Comput Biol. 2019;15 doi: 10.1371/journal.pcbi.1006701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ritchie ME, Phipson B, Wu D. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Buske FA, Bauer DC, Mattick JS, Bailey TL. Triplexator: detecting nucleic acid triple helices in genomic and transcriptomic data. Genome Res. 2012;22:1372–1381. doi: 10.1101/gr.130237.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li JCA. Modeling survival data: extending the cox model. Sociol Method Res. 2003;32:117–120. [Google Scholar]
- 28.Friedman J, Hastie T, Tibshirani R. Regularization paths for generalized linear models via coordinate descent. J Stat Softw. 2010;33:1–22. [PMC free article] [PubMed] [Google Scholar]
- 29.Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shannon P, Markiel A, Ozier O. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ginestet C. ggplot2: elegant graphics for data analysis. J R Stat Soc a Stat. 2011;174 245- [Google Scholar]
- 32.Gu Z, Eils R, Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016;32:2847–2849. doi: 10.1093/bioinformatics/btw313. [DOI] [PubMed] [Google Scholar]
- 33.Gagliardi M, Matarazzo MR. RIP: RNA immunoprecipitation. Methods Mol Biol. 2016;1480:73–86. doi: 10.1007/978-1-4939-6380-5_7. [DOI] [PubMed] [Google Scholar]
- 34.Filtz TM, Vogel WK, Leid M. Regulation of transcription factor activity by interconnected post-translational modifications. Trends Pharmacol Sci. 2014;35:76–85. doi: 10.1016/j.tips.2013.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Francois M, Donovan P, Fontaine F. Modulating transcription factor activity: Interfering with protein-protein interaction networks. Semin Cell Dev Biol. 2018 doi: 10.1016/j.semcdb.2018.07.019. [DOI] [PubMed] [Google Scholar]
- 36.Gao F, Foat BC, Bussemaker HJ. Defining transcriptional networks through integrative modeling of mRNA expression and transcription factor binding data. BMC Bioinformatics. 2004;5:31. doi: 10.1186/1471-2105-5-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hainaut P, Hollstein M. p53 and human cancer: the first ten thousand mutations. Adv Cancer Res. 2000;77:81–137. doi: 10.1016/s0065-230x(08)60785-x. [DOI] [PubMed] [Google Scholar]
- 38.Goh AM, Coffill CR, Lane DP. The role of mutant p53 in human cancer. J Pathol. 2011;223:116–126. doi: 10.1002/path.2784. [DOI] [PubMed] [Google Scholar]
- 39.Mohler JL, Antonarakis ES, Armstrong AJ. Prostate cancer, Version 2.2019, NCCN clinical practice guidelines in oncology. J Natl Compr Canc Netw. 2019;17:479–505. doi: 10.6004/jnccn.2019.0023. [DOI] [PubMed] [Google Scholar]
- 40.Chatterjee P, Schweizer MT, Lucas JM. Supraphysiological androgens suppress prostate cancer growth through androgen receptor-mediated DNA damage. J Clin Invest. 2019;130:4245–4260. doi: 10.1172/JCI127613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cascon A, Robledo M. MAX and MYC: a heritable breakup. Cancer Res. 2012;72:3119–3124. doi: 10.1158/0008-5472.CAN-11-3891. [DOI] [PubMed] [Google Scholar]
- 42.Shen N, Zhao J, Schipper JL. Divergence in DNA specificity among paralogous transcription factors contributes to their differential in vivo binding. Cell Syst. 2018;6:470-83 e8. doi: 10.1016/j.cels.2018.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Han ME, Baek SJ, Kim SY, Kang CD, Oh SO. ATOH1 can regulate the tumorigenicity of gastric cancer cells by inducing the differentiation of cancer stem cells. PLoS One. 2015;10 doi: 10.1371/journal.pone.0126085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Klisch TJ, Vainshtein A, Patel AJ, Zoghbi HY. Jak2-mediated phosphorylation of Atoh1 is critical for medulloblastoma growth. Elife. 2017;6:e31181. doi: 10.7554/eLife.31181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Klinge CM. Non-coding RNAs: long non-coding RNAs and microRNAs in endocrine-related cancers. Endocr Relat Cancer. 2018;25:R259–RR82. doi: 10.1530/ERC-17-0548. [DOI] [PubMed] [Google Scholar]
- 46.Li Y, Syed J, Sugiyama H. RNA-DNA triplex formation by long noncoding RNAs. Cell Chem Biol. 2016;23:1325–1333. doi: 10.1016/j.chembiol.2016.09.011. [DOI] [PubMed] [Google Scholar]
- 47.Mas-Ponte D, Carlevaro-Fita J, Palumbo E, Hermoso Pulido T, Guigo R, Johnson R. LncATLAS database for subcellular localization of long noncoding RNAs. RNA. 2017;23:1080–1087. doi: 10.1261/rna.060814.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang T, Tan P, Wang L. RNALocate: a resource for RNA subcellular localizations. Nucleic Acids Res. 2017;45:D135–D1D8. doi: 10.1093/nar/gkw728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xu S, Wang T, Song W. The inhibitory effects of AR/miR-190a/YB-1 negative feedback loop on prostate cancer and underlying mechanism. Sci Rep. 2015;5:13528. doi: 10.1038/srep13528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Alon U. CRC; Boca Raton, FL: 2007. An introduction to systems biology: design principles of biological circuits. [Google Scholar]
- 51.Dhingra P, Martinez-Fundichely A, Berger A. Identification of novel prostate cancer drivers using RegNetDriver: a framework for integration of genetic and epigenetic alterations with tissue-specific regulatory network. Genome Biol. 2017;18:141. doi: 10.1186/s13059-017-1266-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Roy N, Hebrok M. Regulation of cellular identity in cancer. Dev Cell. 2015;35:674–684. doi: 10.1016/j.devcel.2015.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sheng K, Lu J, Zhao H. ELK1-induced upregulation of lncRNA HOXA10-AS promotes lung adenocarcinoma progression by increasing Wnt/beta-catenin signaling. Biochem Biophys Res Commun. 2018;501:612–618. doi: 10.1016/j.bbrc.2018.04.224. [DOI] [PubMed] [Google Scholar]
- 54.Al-Kershi S, Bhayadia R, Ng M. The stem cell-specific long noncoding RNA HOXA10-AS in the pathogenesis of KMT2A-rearranged leukemia. Blood Adv. 2019;3:4252–4263. doi: 10.1182/bloodadvances.2019032029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Szklarczyk D, Gable AL, Lyon D. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607–DD13. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.