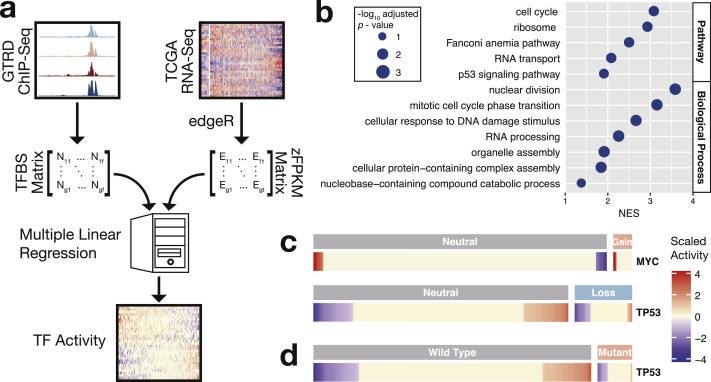
Fig. 1.
Calculation of transcription factor activity in normal prostates and primary prostate cancers. (a) Computational workflow of transcription factor activity. In brief, multiple linear regression was applied to the zFPKM values of a gene set and binding site numbers of transcription factors in the proximal regulatory regions of the same gene set. The selected set of genes were those differentially expressed between tumors and normal tissues. Regression coefficients estimated transcription factor activity. (b) Biological roles of the genes showing expression correlation with MYC activity (BH adjusted p-value < 0.05), which were in accordance with the reported functions of MYC. (c) Genomic copy number variations and (d) mutations affected TP53 and MYC activity. Mutations included deleterious mutations, missense, and in-frame INDELs. zFPKM, standardized fragments per kilobase per million mapped fragments.