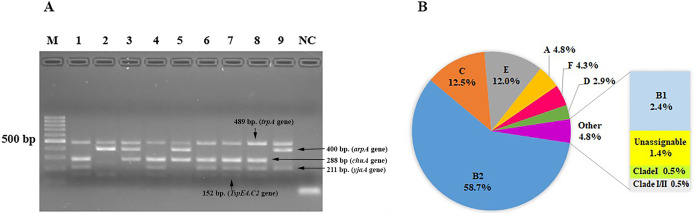
Figure 1. The distribution of phylogenetic groups among uropathogenic Escherichia coli isolates by the new Clermont phylo-typing method.
(A) Multiplex PCR profiles for specific uropathogenic Escherichia coli isolates by detecting the arpA (400 bp), chuA (288 bp), yjaA (211 bp), and TspE4.C2 (152 bp) genes. Lane M, 100-base pair ladder (Fermantas); Lane 1, group B2 (-, +, +, +); Lane 2, group B1 (+, -, -, +); Lane 3, group D or E (+, +, -,-); Lane 4, group B2 (-, +, +, +); Lane 5, group D or E (+, +, -,-); Lane 6, group B2 (-, +, +, +); Lane 7, group B2 (-, +, +, +); Lane 8, group B2 (-, +, +, +); Lane 9, group A or C (+, -, +, -); Lane NC, negative control. The trpA (489 bp) internal control gene appeared in all samples except the negative control. Distilled water without any DNA as negative controls was used in PCR experiments. (B) The percentage of phylogenetic groups among uropathogenic Escherichia coli isolates.