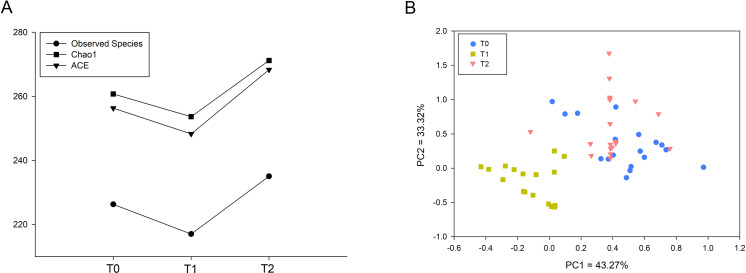
Figure 1. Comparison of microbial diversity of T0, T1 and T2.
(A) Sequences (60,518) were randomly subsampled to obtain equal numbers of sequences from each dataset. Indices, namely, Observed Species, Chao1 and ACE, representing α diversity are shown. (B) Phylogenetic distances between samples were calculated via Bray–Curtis distance. Each dot in the scatter plot represents one sample. The percentage of variation is indicated on the x and y-axes. T1 significantly differed from T0 and T2 (ANOSIM, p < 0.05).