Abstract
Three-dimensional (3D) in vitro culture models better recapitulate the tissue microenvironment, and therefore may provide a better platform to evaluate therapeutic effects on adhesive cell–cell interactions. The objective of this study was to determine if AD-01, a peptide derivative of FK506-binding protein like that is reported to bind to the adhesion receptor CD44, would induce a greater reduction in breast epithelial spheroid adhesion to endothelial tube-like networks in our 3D coculture model system compared to two-dimensional (2D) culture. MCF10A, MCF10A-NeuN, MDA-MB-231, and MCF7 breast epithelial cells were pretreated with AD-01 either as single cells or as spheroids. Breast epithelial cell adhesion to 2D tissue culture substrates was first measured, followed by spheroid formation (breast cell–cell adhesion) and spheroid adhesion to Matrigel or endothelial networks. Finally, CD44 expression was quantified in breast epithelial cells in 2D and 3D culture. Our results show that AD-01 had the largest effect on spheroid formation, specifically in breast cancer cell lines. AD-01 also inhibited breast cancer spheroid adhesion to and migration along endothelial networks. The different breast epithelial cell lines expressed more CD44 when cultured as 3D spheroids, but this did not universally translate into higher protein levels. This study shows that 3D coculture models can enable unique insights into cell adhesion, migration, and cell–cell interactions, thereby enhancing understanding of basic biological mechanisms. Furthermore, such 3D coculture systems may also represent a more relevant testing platform for understanding the mechanism-of-action of new therapeutic agents.
Impact Statement
Cell adhesion is inherently different in two dimensional (2D) compared to three dimensional (3D) culture; yet, most adhesion assays in academia and industry are still conducted in 2D because few simple, yet effective, adhesion models exist in 3D. Recently we developed a 3D in vitro coculture model to examine breast epithelial spheroid interactions with endothelial tubes. We now show that this 3D coculture model can effectively be used to interrogate and quantify drug-induced differences in breast epithelial cell adhesion that are unique to 3D cocultures. This 3D coculture adhesion model can furthermore be modified for use with other cell types to better predict drug effects on cell-vasculature adhesion.
Keywords: cell adhesion, breast spheroids, 3D culture models, endothelial cells, CD44
Introduction
In vitro cell-based assays are widely used to study cancer therapies by quantifying cell characteristics, including proliferation, adhesion, and migration. However, conventional two-dimensional (2D) cell culture fails to fully capture the complex biomechanical and biochemical cues in the tumor microenvironment, often resulting in ineffective estimates of therapeutic potential.1–4 Three-dimensional (3D) human cancer models, such as breast epithelial spheroids, better recapitulate many in vivo cell functions, including increased chemoresistance.4–8 More recently, 3D culture models were designed to also include other cell types through direct contact9,10 or Transwell plates.11,12 These 3D coculture models enhance the cell–cell interactions that are necessary to study adhesion, migration, and tumor invasion.7,13–15 We recently created a 3D coculture model system of endothelial cell tube-like networks and breast epithelial spheroids, which demonstrated that breast epithelial spheroids preferentially adhered to and migrated along the endothelial networks.9
CD44 is a cell surface adhesion molecule that is upregulated in breast cancer and is gaining importance in identifying cancer stem cells and as a cancer diagnostic marker.16,17 CD44 expression is thought to negatively correlate with breast cancer disease-free survival in human patients and, in a mouse model, CD44 knockdown reduced MDA-MB-231 tumor burden and increased survival.18 CD44 is also reported to mediate cancer cell adhesion to endothelial cells.19 CD44 has many different isoforms due to splice variants, but each CD44 isoform binds to hyaluronic acid, which is present both in the extracellular matrix and on the endothelial surface.20 Fontana et al. recently showed increased CD44 expression in PC3 prostate cancer cells in 3D compared to 2D culture.21 However, they did not measure 2D versus 3D CD44-mediated cancer cell adhesion, despite evidence that cell–cell and cell–matrix adhesion are inherently different in 3D.22,23
FK506-binding protein like (FKBPL) is an immunophilin protein family member that is emerging as a potential biomarker for breast cancer.24,25 FKBPL and its peptide derivative AD-01 have been reported to bind to CD44, reducing endothelial cell in vitro migration and in vivo angiogenesis.26,27 FKBPL and AD-01 also reduced breast cancer spheroid formation, likely by reducing the CD44+ cancer stem cell population.28 While FKBPL and AD-01 have been evaluated in endothelial and breast cancer cell lines separately, their effects on cell adhesion have not yet been studied in a 3D coculture model.
In this study, our goal was to determine if AD-01 would decrease breast epithelial spheroid adhesion to endothelial cell tube-like networks in our complex 3D coculture model, and if the adhesion effect would be greater in 3D culture due to increased CD44 expression. We therefore assessed breast cancer cell viability and adhesion in 2D and 3D monoculture and coculture with endothelial tube-like networks. Finally, we quantified CD44 expression and protein levels in breast epithelial cell lines in both 2D and 3D culture.
Materials and Methods
Cell culture and reagents
Breast epithelial cells were provided by Mauricio Reginato (Drexel College of Medicine). The MCF10A breast epithelial cell line, which is nontumorigenic and does not express estrogen receptor,29 was cultured in Dulbecco's Modified Eagle Medium (DMEM)/F12 (Corning) supplemented with 5% horse serum (Invitrogen), 20 ng/mL epidermal growth factor (EGF; Peprotech), 10 μg/mL bovine insulin (Sigma), 10 ng/mL cholera toxin (Enzo), 500 ng/mL hydrocortisone (Sigma), and 1% penicillin–streptomycin (Invitrogen). MCF10A cells that overexpressed EGF receptor 2 (ERBB2, HER2/neu; abbreviated as MCF10A-NeuN), which is prevalent in breast cancer and portends a poor outcome,30 were cultured in the same medium as MCF10A cells. The triple-negative MDA-MB-231 breast cancer cell line31 was cultured in DMEM (Corning) with 10% fetal bovine serum (FBS; Hyclone) and 1% penicillin–streptomycin. The estrogen and progesterone receptor-positive MCF-7 cell line32 was cultured in MDA-MB-231 cell medium with added 10 μg/mL bovine insulin (Sigma). Human umbilical vein endothelial cells (HUVECs; Cell Applications) were cultured in endothelial growth medium-2 (Lonza) supplemented with 10% FBS and 1% penicillin–streptomycin. For some experiments, MCF10A and MCF10A-NeuN cells were stably transfected with green fluorescent protein (Takara Bio, Mountain View, CA). Cells were maintained in an incubator at 37°C and 5% CO2 and given fresh medium every 2 days. Breast cells were cultured up to passage 25, while HUVECs were cultured to passage 7.
AD-01, a synthetic peptide derived from the human FKBPL protein, was provided by ALMAC Discovery for research purposes.26,27,33 AD-01 was obtained as a lyophilized powder, dissolved in phosphate-buffered saline (PBS, pH 7.4), and stored at −20°C.
Formation of breast cell 3D spheroids
We formed breast cell 3D spheroids using an established protocol.9,34 Briefly, we added 30 μL undiluted Matrigel (Corning; growth factor reduced) to each well of a BioCoat Falcon Culture slide (Corning), which was then gelled for 20 min at 37°C. Approximately, 5000 breast epithelial cells were seeded in each well in the appropriate culture medium with 20% added Matrigel. Spheroids were provided with fresh medium every 4 days. MCF10A cells took 8–10 days to form spheroids, whereas MCF10A-NeuN, MDA-MB-231, and MCF-7 cells took 5–6 days to form spheroids. We defined MCF10A and MCF10A-NeuN cells as spheroids when they organized into polarized acini of an epithelial cell ring surrounding a hollow lumen. MDA-MB-231 and MCF-7 spheroids were less organized and were instead defined by their size. Average spheroid sizes were ∼80 μm for MCF10A, 120 μm for MCF10A-NeuN, 100 μm for MDA-MB-231, and 100 μm for MCF-7 cells.
Formation of endothelial cell tube-like networks
Endothelial tube-like networks were formed by adding Matrigel into each well of a BioCoat Falcon Culture slide as described previously for the spheroids. Approximately, 100,000 HUVEC were first labeled with CellTracker Red (1:1000; Invitrogen) and then added to each well in 200 μL serum-free endothelial basal medium-2 (Lonza). Samples were maintained at 37°C, 5% CO2 for 6 h to form networks. Endothelial network characteristics were quantified using the Angiogenesis Analyzer for ImageJ. Junctions were defined as pixels with at least three neighbors; segments were defined as elements delimited by two junctions; and branches were defined as elements delimited by a junction and one extremity.
Confocal microscopy
Cells and spheroids were fixed with 4% paraformaldehyde for 1 h. Samples were then incubated with a 90 min primary block of 10% goat serum in an immunofluorescence (IF) buffer (130 mM NaCl, 7 mM Na2HPO4, 3.5 mM NaH2PO4, 7.7 mM NaN3, 0.1% bovine serum albumin [BSA; Sigma], 0.2% Triton X-100, and 0.05% Tween-20, pH 7.4) followed by a 40 min secondary block of 10% goat serum in IF buffer with Affinipure F(ab′)2 fragment goat anti-mouse immunoglobulin G (115-006-020, 1:100; Jackson ImmunoResearch). Samples were then labeled using an integrin α6 primary antibody (MAB1378, 1:100 overnight at 4°C; Millipore) and an Alexa Fluor 488 secondary antibody (A-11001, 1:200; Invitrogen), and bisbenzimide (nuclei, 1:1000 for 1 h at room temperature; ThermoScientific). Samples were washed and mounted in Prolong Gold Antifade (P36934; ThermoScientific). HUVEC networks prelabeled with CellTracker Red were fixed with 4% paraformaldehyde, labeled for nuclei, and mounted as described. We then imaged the cells and spheroids by confocal microscopy using 5–8 slice z stacks (2 μm steps). In some cases, images were combined into a 4 × 4 tile as a stitched extended focus (compressed) with a 10% overlap using Volocity 6.3 (Perkin Elmer).
Cell viability
We assessed cell viability with a Live/Dead Assay (Invitrogen) as per manufacturer's instructions. Briefly, live cells were identified using calcein-AM (0.5 μM), while dead cells were identified using ethidium bromide (1 μM). Samples were incubated for 20 min at room temperature and then imaged by confocal microscopy as described. Live versus dead cells were quantified using the Cell counter plugin in Image J.
Adhesion assays
For 2D cell adhesion, trypsinized endothelial and breast epithelial cells were pretreated with AD-01 for 30 min with gentle rocking and then added onto tissue culture dishes that were uncoated, coated with 20 μg/mL collagen type I (rat tail), or coated with 50 μg/mL Matrigel. After 24 h, unadhered cells were removed by gentle washing. Cells were fixed in 4% paraformaldehyde and labeled with bisbenzimide (1:1000). Samples were then imaged by confocal microscopy as a 3 × 3 tile, with two z slices per image (step size 0.5 μm). At least three images were taken for each sample. Adhered cells were quantified by counting nuclei using the Cell counter plugin in Image J.
For 3D cell adhesion, preformed spheroids or HUVEC networks were treated with AD-01 for 24 and 48 h. After gentle washing, samples were fixed, labeled, and imaged as described. Spheroid number and diameter were quantified using the Measure ImageJ plugin. In addition, breast epithelial cells were pretreated with AD-01 for 1 h with gentle rocking and then seeded for spheroids. Spheroid number and diameter were quantified after 8 days. Finally, for 3D coculture adhesion, preformed breast spheroids were pipetted onto HUVEC networks. AD-01 was added to the coculture 2 h after adding spheroids. The coculture was then maintained for 24 h. After gentle washing, samples were fixed, labeled, imaged, and quantified as described.
Reverse transcriptase-polymerase chain reaction
CD44 expression was measured using SYBR green quantitative reverse transcriptase-polymerase chain reaction. Spheroids were first recovered from Matrigel using Cell Recovery Solution (VWR). Total RNA from cells and spheroids was isolated using Trizol (Invitrogen) and quantified using a Nanodrop Spectrophotometer (ThermoFisher). Two micrograms RNA per sample was combined with 10 × buffer, deoxynucleoside triphosphates, random primers, RNAse inhibitor, and reverse transcriptase in nuclease-free water. cDNA master mix was then added, and samples run in a thermocycler (Eppendorf) for 10 min at 25°C, 120 min at 37°C, and 5 min at 85°C, and then cooled to 4°C. For quantitative PCR, 200 ng of cDNA and SYBR green was mixed with the appropriate primers (Table 1) in nuclease-free water and amplified using an Eppendorf Mastercycler for 2 min at 95°C, and 40 cycles of 15 s at 95°C, 15 s at 55°C, and 20 s at 60°C. ΔCt was used to calculate gene expression as a fold change of experimental Ct value compared to β-actin.35
Table 1.
Human CD44s, CD44v2-CD44v10, and β-Actin Polymerase Chain Reaction Primer Sequences
| Forward primer | Reverse primer | |
|---|---|---|
| CD44s | GGAGCAGCACTTCAGGAGGTTAC | GGAATGTGTCTTGGTCTCTGGTAGC |
| CD44v2 | ATCACCGACAGCACAGACAGAAT | AACCATGAAAACCAATCCCAGG |
| CD44v3 | TACGTCTTCAAATACCATCTCAGCA | AATCTTCATCATCATCAATGCCTG |
| CD44v4 | AACCACACCACGGGCTTTTG | TCCTTGTGGTTGTCTGAAGTAGCA |
| CD44v5 | TGCTTATGAAGGAAACTGGAAC | TGTGCTTGTAGAATGTGGGGT |
| CD44v6 | CCAGGCAACTCCTAGTAGTACAACG | CGAATGGGAGTCTTCTTTGGGT |
| CD44v7 | GCCTCAGCTCATACCAGCCATC | TCCTTCTTCCTGCTTGATGACCT |
| CD44v8 | TGGACTCCAGTCATAGTATAACGC | GGTCCTGTCCTGTCCAAATC |
| CD44v9 | AGCAGAGTAATTCTCAGAGC | TGATGTCAGAGTAGAAGTTGTT |
| CD44v10 | CCTCTCATTACCCACACACG | CAGTAACTCCAAAGGACCCA |
| β-actin | GTGAAGGTGACAGCAGTCGGTT | GAAGTGGGGTGGCTTTTAGGAT |
Western blot
CD44 protein levels were determined by Western blot. Cells were scraped in lysis buffer (20 mM Tris, 150 mM NaCl, 1% Triton X-100, 2 mM ethylenediaminetetraaceticacid, 2 mM phenylmethylsulfonyl fluoride, 0.1% sodium dodecyl sulfate [SDS], 2 mM Na3VO4, 50 mM NaF, 10% glycerol, and complete protease inhibitor [Roche], pH 7.4). Insoluble material was removed by spinning samples for 10 min at 10,000 g. After normalizing cell lysates for protein content using a BCA Assay (ThermoFisher), samples were separated by SDS-polyacrylamide gel electrophoresis on a 4–12% Bis-Tris gel (ThermoFisher), transferred to a nitrocellulose membrane (ThermoFisher), and blocked using 5% BSA. Membranes were labeled using a CD44 (v3-v10) primary antibody (ab119365, 1:1000; Abcam) overnight followed by a secondary antibody (Promega; 1:2000) for 1 h. The loading control was β-actin (SC47778-C4; Santa Cruz). Membranes were then incubated with an enhanced chemiluminescence kit (Western Lightning; PerkinElmer), protein bands were imaged with a Fluorchem digital imager (Alpha Innotech), and protein band intensity was quantified using AlphaEase FC software.
Statistical analysis
GraphPad Prism was used for all statistical analyses. Student's t-test was used to compare two groups, while analysis of variance with Tukey–Kramer posthoc test was used to compare multiple groups. Data are shown as mean ± standard deviation. All experiments were conducted with three samples per experiment and repeated at least three times.
Results
AD-01 did not affect 2D or 3D breast epithelial cell viability
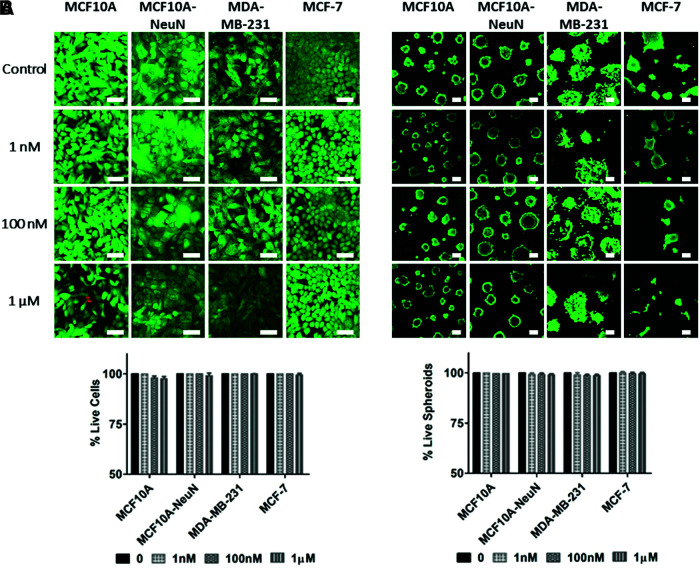
Although AD-01 has been studied in 2D cell migration and adhesion,26–28 its efficacy had not been evaluated in 3D breast epithelial spheroids cocultured with endothelial tubes. We first measured whether AD-01 would impact breast epithelial cell viability in 2D and 3D culture (Fig. 1). When increasing AD-01 concentrations were added to breast epithelial cells cultured on 2D uncoated tissue culture polystyrene, breast epithelial cells remained ∼98% viable even at the highest AD-01 concentration. Similarly, 3D breast epithelial spheroids cultured in Matrigel were more than 97% viable at the highest peptide concentration. Both MCF10A and MCF10A-NeuN cell lines formed polarized acini with hollow central lumens, as indicated by the lack of Live/Dead staining at the spheroid center. These data show that AD-01 did not affect 2D or 3D breast epithelial cell viability and is not cytotoxic.
FIG. 1.
AD-01 did not affect breast epithelial cell viability, whether the cells were cultured as 2D monolayers or as 3D spheroids. (A) Breast epithelial monolayers on uncoated tissue culture polystyrene and (B) breast epithelial spheroids on Matrigel were treated with increasing AD-01 concentrations (0, 1 nM, 100 nM, and 1 μM) for 48 h and then labeled with Calcein AM (live cells, green) and ethidium homodimer (dead cells, red), and imaged by confocal microscopy. The % of live cells was then quantified using ImageJ and normalized to the untreated control (0 nM AD-01). Scale bar = 50 μm. 2D, two dimensional; 3D, three dimensional. Color images are available online.
AD-01 decreased 2D and 3D breast epithelial cell adhesion
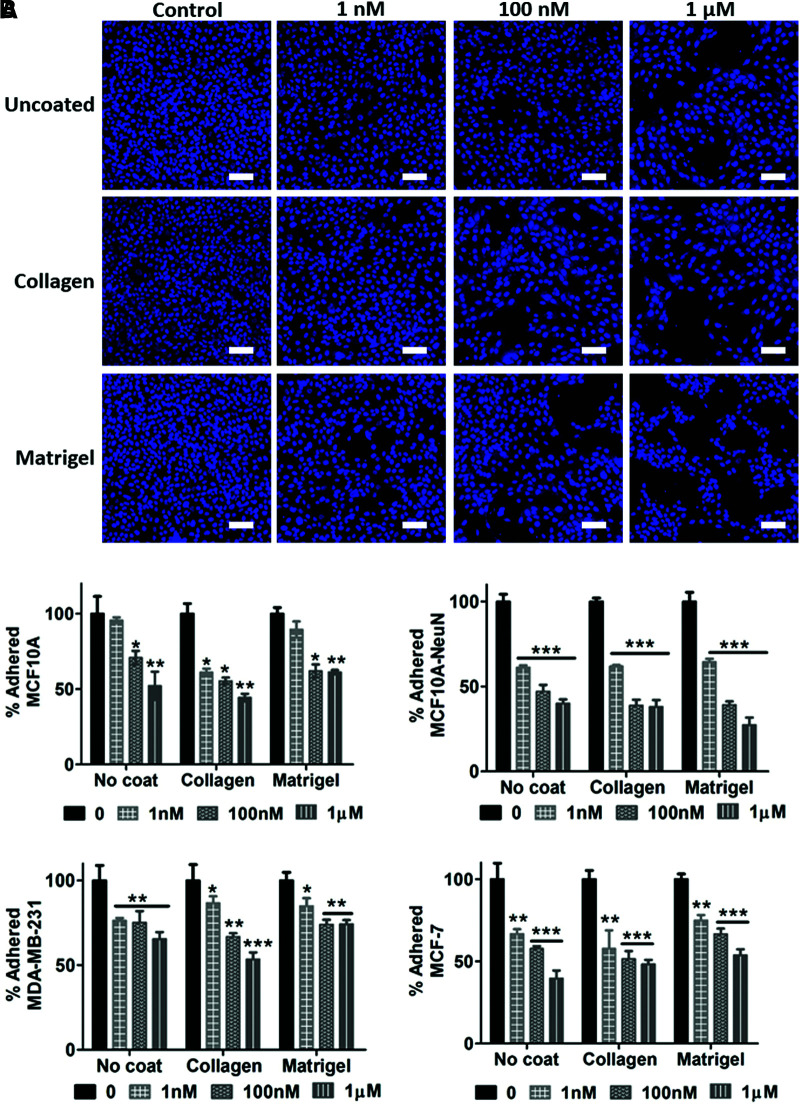
We then determined whether AD-01 affected breast epithelial cell adhesion in 2D and 3D. Adhered 2D breast epithelial cells decreased in number when pretreated with increasing AD-01 concentrations on uncoated, collagen-coated, or Matrigel-coated substrates (Fig. 2). The decrease in cell adhesion was statistically significant when compared to the untreated control (0 nM AD-01) for each cell type and substrate coating, with the exception of MCF10A cells on uncoated and Matrigel-coated surfaces at 1 nM AD-01. Overall, adhered MCF10A-NeuN cells decreased the most (up to 73% at 1 μM AD-01 on Matrigel), while adhered MDA-MB-231 cells decreased the least (only 26% decrease at 1 μM AD-01 on Matrigel). There was no statistically significant difference in cell adhesion among the three substrate coatings.
FIG. 2.
AD-01 pretreatment decreased 2D breast epithelial cell adhesion. Suspended breast epithelial cells were pretreated with increasing AD-01 concentrations for 30 min, after which cells were seeded on uncoated tissue culture polystyrene (uncoated), or the same dishes coated with 10 μg/mL collagen or 50 μg/mL Matrigel. After 24 h, samples were then fixed, labeled for nuclei (bisbenzimide, blue), imaged by confocal microscopy, and quantified using ImageJ. (A) Representative images of MCF10A-NeuN breast epithelial cells pretreated with increasing AD-01 concentrations; and (B) quantification of attached cells after 24 h, normalized to untreated samples for each cell type substrate. *p < 0.01, **p < 0.001, ***p < 0.0001. Scale bar = 100 μm. Color images are available online.
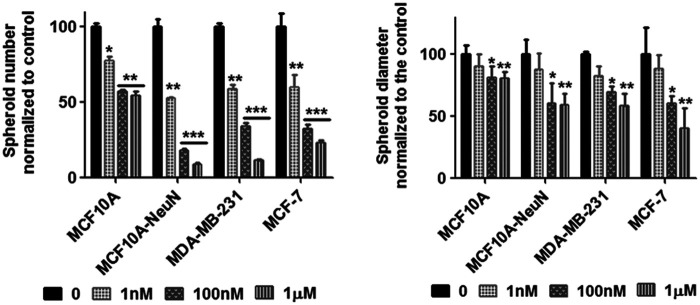
We next studied whether this decreased breast epithelial cell adhesion with AD-01 impacted spheroid formation (Fig. 3). Breast epithelial cells were considered spheroids if they were greater than 50 μm in size. In addition, MCF10A and MCF10A-NeuN cells were only considered spheroids if they were organized into polarized acini with a hollow lumen surrounded by a single ring of epithelial cells. Breast epithelial cells that were pretreated with AD-01 successfully formed spheroids; however, the resulting spheroids were statistically significantly smaller in size and fewer in number. MCF10A spheroid number was the least affected, decreasing by around 50% with 1 μM AD-01. In contrast, MCF10A-NeuN, MDA-MB-231, and MCF-7 spheroid number decreased by 90%, 88.5%, and 76.6% respectively, at 1 μM AD-01. Single cells were even observed in the MCF10A-NeuN and MCF-7 3D cultures at the highest AD-01 concentration. Similarly, MCF10A spheroid diameter decreased by only 20% at 1 μM AD-01, whereas MCF10A-NeuN, MDA-MB-231, and MCF-7 spheroid diameter decreased by 40–60%.
FIG. 3.
AD-01 pretreatment of single cells decreased the number and diameter of breast epithelial spheroids. Suspended breast epithelial cells were treated with AD-01 and then seeded to form spheroids. After 5–8 days, spheroids were fixed, labeled for integrin α6 (MCF10A and MCF10A-NeuN) or F-actin (MDA-MB-231 and MCF-7) and nuclei, and imaged by confocal microscopy. Attached spheroid number and diameter were determined using ImageJ. *p < 0.01, **p < 0.001, ***p < 0.0001 compared to untreated spheroids of each cell type (0 nM).
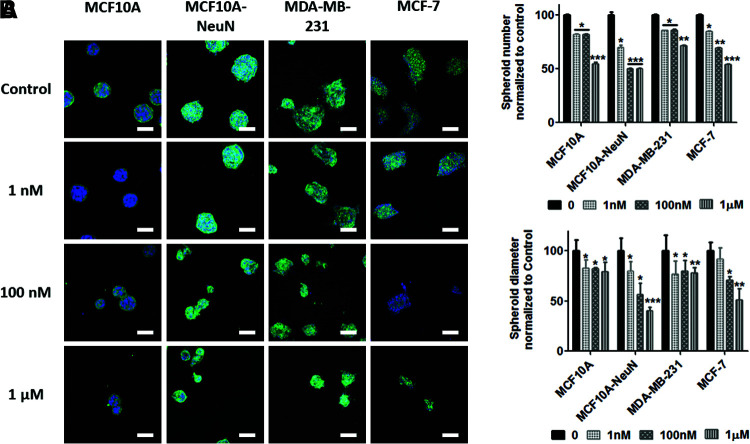
We next treated preformed 3D breast epithelial spheroids with AD-01 to determine if the peptide affected their adhesion. AD-01 treatment again significantly decreased the number of spheroids that remained attached as well as the spheroid diameter (Fig. 4). Spheroid number decreased by 45–50% at 1 μM AD-01, with MDA-MB-231 breast epithelial spheroids the least affected (29%). Spheroid diameter decreased by more than 50% in MCF10A-NeuN and MCF-7 spheroids treated with 1 μM AD-01, whereas spheroid diameter only decreased by 25% in MCF10A and MDA-MB-231 treated with 1 μM AD-01.
FIG. 4.
AD-01 treatment decreased the number and diameter of preformed spheroids. (A) Preformed breast epithelial spheroids were treated with increasing AD-01 concentrations for 48 h and then labeled for integrin α6 (MCF10A and MCF10A-NeuN: green) or F-actin (MDA-MB-231 and MCF-7: green) and nuclei (bisbenzimide: blue). Samples were then imaged by confocal microscopy. Scale bar = 50 μm. (B) Quantification of spheroid number and diameter using ImageJ. *p < 0.01; **p < 0.001; ***p < 0.0001 compared to the untreated spheroids of each cell type (0 nM). Color images are available online.
Breast 3D spheroids did not migrate along endothelial tubes with AD-01
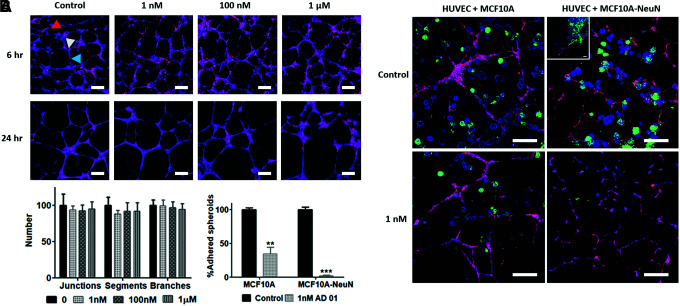
We previously observed that breast 3D spheroids attached to and migrated along HUVEC tube-like networks in coculture.9 We therefore investigated the AD-01 effect on breast spheroid adhesion to HUVEC networks. AD-01 did not affect HUVEC tube-like networks over 48 h of treatment (Fig. 5A). The number of endothelial network junctions, segments, and branches was statistically similar among all AD-01 concentrations. When preformed breast epithelial spheroids were added to preformed HUVEC networks, both MCF10A and MCF10A-NeuN breast epithelial spheroids attached to the endothelial networks after 24 h of coculture (Fig. 5B). MCF10A-NeuN cells also migrated along the endothelial cells (Fig. 5B, inset). However, when the 3D coculture was treated with 1 nM AD-01, the number of adhered MCF10A spheroids decreased by more than 50% (p < 0.001), and continued to decrease at higher AD-01 concentrations (data not shown). AD-01 reduced MDA-MB-231 and MCF-7 spheroid adhesion to endothelial networks in a manner similar to MCF10A spheroids. In contrast, 1 nM AD-01 nearly eliminated MCF10A-NeuN spheroid adhesion to endothelial networks (p < 0.0001). The few MCF10A-NeuN spheroids that attached to the endothelial networks did not elongate along the networks.
FIG. 5.
While AD-01 did not affect HUVEC network formation or adhesion, AD-01 treatment decreased MCF10A-NeuN adhesion to and migration along HUVEC tube-like networks. (A) Confocal microscopy images of HUVEC networks after 6 and 24 h of 0, 1 nM, 100 nM, or 1 μM AD-01 treatment. HUVECs were labeled with CellTracker (red) and bisbenzimide (nuclei, blue). Quantification of network junctions (white arrowhead), segments (blue arrowhead), and branches (red arrowhead) after 24 h of AD-01 treatment using ImageJ, normalized to the untreated control (0 nM AD-01). Scale bar = 200 μm. (B) Confocal microscopy images of MCF10A and MCF10A-NeuN breast epithelial spheroids cocultured with HUVEC networks. Preformed breast spheroids from green fluorescent protein-expressing cells (green) were pipetted onto preformed HUVEC networks (CellTracker, red) and treated with 1 nM AD-01 after 2 h of coculture. Samples were also labeled with bisbenzimide (nuclei, blue). Quantification of adhered spheroids as a % of control. Scale bar = 200 μm. Inset shows MCF10A-NeuN cells migrating along endothelial networks. Scale bar = 50 μm. **p < 0.001, ***p < 0.0001. HUVEC, human umbilical vein endothelial cell. Color images are available online.
AD-01 adhesion effects may relate to CD44 gene expression
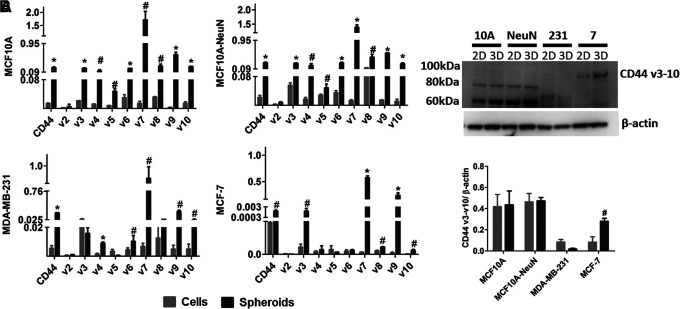
In prior studies, AD-01 was reported to reduce 2D cell adhesion and migration of breast (MCF-7 and MDA-MB-231) and pancreatic cells (PC-3) by binding to CD44.26,27 We therefore measured CD44 gene expression and protein levels to determine if CD44 might explain the variation among breast epithelial cell lines and difference in 2D versus 3D culture (Fig. 6). CD44 mRNA was highest and quite similar in MCF10A and MCF10A-NeuN cells. CD44 mRNA was slightly lower in MDA-MB-231 cells, and lower still in MCF-7 cells. In all breast epithelial cell types, CD44s mRNA and its variants were expressed more highly in 3D spheroids than in 2D monolayers. CD44 protein levels did not vary between 2D and 3D cultures, except for MCF-7, which showed significantly higher protein expression in 3D spheroids versus a 2D monolayer. While MCF10A and MCF10A-NeuN cells showed similar CD44 molecular weight distributions, the primary CD44 isoform in MDA-MB-231 cells was ∼70 kDa, while the primary CD44 isoform in MCF-7 was ∼90 kDa.
FIG. 6.
CD44 expression was significantly higher in 3D breast epithelial spheroids compared to 2D monolayers for all cell types, whereas CD44 protein only increased in 3D MCF-7 spheroids. (A) Reverse transcriptase-polymerase chain reaction for CD44s and v2-v10 isoforms for 2D breast epithelial cells and 3D breast epithelial spheroids. (B) Western blot with quantification of CD44 v3-v10 protein for MCF10A, MCF10A-NeuN, MDA-MB-231, and MCF-7 breast epithelial cells. #p < 0.05, *p < 0.01.
Discussion
Three-dimensional cell culture platforms may provide a more appropriate model for evaluating cell adhesion mechanisms, as they better capture the complex cell–cell and cell–matrix interactions in the tissue microenvironment observed in vivo. We now show that AD-01, which reportedly binds to CD44, reduced breast epithelial cell adhesion in both 2D and 3D, but had the largest effect on breast spheroid formation in MCF10A-NeuN and MDA-MB-231 cells. While AD-01 did not appear to affect endothelial cell tube-like networks per se, it decreased breast epithelial spheroid adhesion to and migration along these endothelial networks. This effect was greatest in MCF10A-NeuN cells, which overexpress ERBB2 (HER2/neu), compared to MCF10A cells. Some of the differences observed among the breast epithelial cell lines may additionally relate to CD44 isoform expression.
The 3D adhesion model reveals important differences in breast epithelial cell adhesion. While effects on 2D cell adhesion were largely similar with AD-01 treatment, the effect of AD-01 on 3D spheroid formation and adhesion varied depending on breast epithelial cell type. AD-01 effects have been attributed to interactions with CD44, which mediates adhesion to neighboring cells as well as to the extracellular matrix.16,36,37 We now show that AD-01 significantly inhibits spheroid formation specifically for the breast cancer cell lines (MCF10A-NeuN, MDA-MB-231, and MCF-7). This suggests that CD44 may be more important for cell–cell adhesion in these cancer cells than in the nonmalignant MCF10A cells, which may rely more on other adhesion molecules.
We also show that AD-01 had a larger effect on MCF10A-NeuN cells, which overexpress the EGF receptor family member, ERBB2 (HER2/neu), compared to noncancerous MCF10A cells, and this effect was increased in 3D culture. In our previous study, we showed that MCF10A-NeuN breast epithelial cells migrated out of spheroids and along HUVEC tubes as soon as they were placed in coculture.9 AD-01 treatment significantly inhibited MCF10A-NeuN spheroid adhesion to and migration along HUVEC tube-like networks more so than for MCF10A spheroids. ERBB2/HER2 is regarded as a poor prognostic indicator for breast cancer survival, and its overexpression drives tumor cell survival, tumor growth, and metastasis.38 CD44 can act as a coreceptor to ERBB receptors and colocalizes with EGF receptor; furthermore, CD44 binding to hyaluronan may affect this interaction.20 In addition, ERBB2/HER2 has also been reported to increase the number of cancer stem cells that are CD44+.39 Thus, AD-01 may act through CD44 to specifically decrease ERBB2/HER2-mediated breast epithelial cell adhesion and/or cancer stem cell number.
Our study further confirms differential expression of CD44 isoforms in different breast cancer cell types, which may impact AD-01 efficacy in each cell type.40 CD44 isoforms may have some functional differences in terms of cell adhesion. For example, CD44 v4 expression correlated with E-selectin-mediated adhesion of breast cancer cells to endothelial cell monolayers; v5 expression associated with invasiveness in vitro; and v8-v10 was implicated with regulating reactive oxygen species production.40,41 MDA-MB-231 and MCF-7 breast cancer cells did have lower CD44 expression overall, and in particular, lower expression of CD44 v4, but this did not appear to directly relate to 2D or 3D adhesion. Additional experiments are needed to determine the relative effects of CD44 isoforms in breast epithelial cell 2D and 3D adhesion.
While CD44 expression significantly increased in 3D breast spheroid culture across nearly all CD44 isoforms, we did not observe the correlative increase in CD44 protein. This could relate to different mRNA transcription and translation and increased CD44 protein in 3D spheroids compared to 2D monolayers. In our studies, only MCF-7 cells showed increased CD44 protein levels in 3D culture, which oddly had the smallest mRNA expression change from 2D to 3D culture. MCF-7 cells showed the largest increase in CD44 v3, v7, and v9, suggesting that these isoforms may have higher correlation between mRNA expression and protein level.
Conclusion
Three-dimensional coculture adhesion assays reveal important differences between 2D and 3D adhesion among breast epithelial and endothelial cells, and thus serve as a more reliable platform for interrogating fundamental biological mechanisms between mixed cell types and for providing a basis to understand and then better predict the therapeutic potential of new anticancer agents in vivo.
Disclosure Statement
A.N.C. is an employee of Almac Discovery Ltd.
Funding Information
No funding was received for this article.
References
- 1. Langhans S.A. Three-dimensional in vitro cell culture models in drug discovery and drug repositioning. Front Pharmacol 9, 6, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Morgan M.M., Johnson B.P., Livingston M.K., et al. Personalized in vitro cancer models to predict therapeutic response: challenges and a framework for improvement. Pharmacol Ther 165, 79, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Antoni D., Burckel H., Josset E., and Noel G. Three-dimensional cell culture: a breakthrough in vivo. Int J Mol Sci 16, 5517, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Edmondson R., Broglie J.J., Adcock A.F., and Yang L. Three-dimensional cell culture systems and their applications in drug discovery and cell-based biosensors. Assay Drug Dev Technol 12, 207, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Duval K., Grover H., Han L.H., et al. Modeling physiological events in 2D vs. 3D cell culture. Physiology (Bethesda) 32, 266, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 6. Kapalczynska M., Kolenda T., Przybyla W., et al. 2D and 3D cell cultures—a comparison of different types of cancer cell cultures. Arch Med Sci 14, 910, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Swaminathan S., Hamid Q., Sun W., and Clyne A.M. Bioprinting of 3D breast epithelial spheroids for human cancer models. Biofabrication 11, 025003, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhao Y., Yao R., Ouyang L., et al. Three-dimensional printing of Hela cells for cervical tumor model in vitro. Biofabrication 6, 035001, 2014 [DOI] [PubMed] [Google Scholar]
- 9. Swaminathan S., Ngo O., Basehore S., and Clyne A.M. Vascular endothelial–breast epithelial cell coculture model created from 3D cell structures. ACS Biomater Sci Eng 3, 2999, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Heneweer M., Muusse M., Dingemans M., et al. Co-culture of primary human mammary fibroblasts and MCF-7 cells as an in vitro breast cancer model. Toxicol Sci 83, 257, 2005 [DOI] [PubMed] [Google Scholar]
- 11. Arrigoni C., Bersini S., Gilardi M., and Moretti M. In vitro co-culture models of breast cancer metastatic progression towards bone. Int J Mol Sci 17, 1405, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Carpenter P.M., Sivadas P., Hua S.S., et al. Migration of breast cancer cell lines in response to pulmonary laminin 332. Cancer Med 6, 220, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Betriu N., and Semino C.E. Development of a 3D co-culture system as a cancer model using a self-assembling peptide scaffold. Gels 4, 65, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jaganathan H., Gage J., Leonard F., et al. Three-dimensional in vitro co-culture model of breast tumor using magnetic levitation. Sci Rep 4, 6468, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Leonard F., and Godin B. 3D in vitro model for breast cancer research using magnetic levitation and bioprinting method. Methods Mol Biol 1406, 239, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Goodison S., Urquidi V., and Tarin D. CD44 cell adhesion molecules. Mol Pathol 52, 189, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zoller M. CD44: can a cancer-initiating cell profit from an abundantly expressed molecule? Nat Rev Cancer 11, 254, 2011 [DOI] [PubMed] [Google Scholar]
- 18. McFarlane S., Coulter J.A., Tibbits P., et al. CD44 increases the efficiency of distant metastasis of breast cancer. Oncotarget 6, 11465, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Draffin J.E., McFarlane S., Hill A., Johnston P.G., and Waugh D.J. CD44 potentiates the adherence of metastatic prostate and breast cancer cells to bone marrow endothelial cells. Cancer Res 64, 5702, 2004 [DOI] [PubMed] [Google Scholar]
- 20. Louderbough J.M., and Schroeder J.A. Understanding the dual nature of CD44 in breast cancer progression. Mol Cancer Res 9, 1573, 2011 [DOI] [PubMed] [Google Scholar]
- 21. Fontana F., Raimondi M., Marzagalli M., et al. Epithelial-to-mesenchymal transition markers and cd44 isoforms are differently expressed in 2D and 3D cell cultures of prostate cancer cells. Cells 8, pii:, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Baker B.M., and Chen C.S. Deconstructing the third dimension: how 3D culture microenvironments alter cellular cues. J Cell Sci 125, 3015, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Khalili A.A., and Ahmad M.R. A review of cell adhesion studies for biomedical and biological applications. Int J Mol Sci 16, 18149, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. McKeen H.D., Byrne C., Jithesh P.V., et al. FKBPL regulates estrogen receptor signaling and determines response to endocrine therapy. Cancer Res 70, 1090, 2010 [DOI] [PubMed] [Google Scholar]
- 25. McKeen H.D., Brennan D.J., Hegarty S., et al. The emerging role of FK506-binding proteins as cancer biomarkers: a focus on FKBPL. Biochem Soc Trans 39, 663, 2011 [DOI] [PubMed] [Google Scholar]
- 26. Valentine A., O'Rourke M., Yakkundi A., et al. FKBPL and peptide derivatives: novel biological agents that inhibit angiogenesis by a CD44-dependent mechanism. Clin Cancer Res 17, 1044, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yakkundi A., McCallum L., O'Kane A., et al. The anti-migratory effects of FKBPL and its peptide derivative, AD-01: regulation of CD44 and the cytoskeletal pathway. PLoS One 8, e55075, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. McClements L., Yakkundi A., Papaspyropoulos A., et al. Targeting treatment-resistant breast cancer stem cells with FKBPL and its peptide derivative, AD-01, via the CD44 pathway. Clin Cancer Res 19, 3881, 2013 [DOI] [PubMed] [Google Scholar]
- 29. Qu Y., Han B., Yu Y., et al. Evaluation of MCF10A as a reliable model for normal human mammary epithelial cells. PLoS One 10, e0131285, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Haenssen K.K., Caldwell S.A., Shahriari K.S., et al. ErbB2 requires integrin alpha5 for anoikis resistance via Src regulation of receptor activity in human mammary epithelial cells. J Cell Sci 123, 1373, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chavez K.J., Garimella S.V., and Lipkowitz S. Triple negative breast cancer cell lines: one tool in the search for better treatment of triple negative breast cancer. Breast Dis 32, 35, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Levenson A.S., and Jordan V.C. MCF-7: the first hormone-responsive breast cancer cell line. Cancer Res 57, 3071, 1997 [PubMed] [Google Scholar]
- 33. Yakkundi A., Bennett R., Hernandez-Negrete I., et al. FKBPL is a critical antiangiogenic regulator of developmental and pathological angiogenesis. Arterioscler Thromb Vasc Biol 35, 845, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Debnath J., Muthuswamy S.K., and Brugge J.S. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods 30, 256, 2003 [DOI] [PubMed] [Google Scholar]
- 35. Li Z., Chen K., Jiang P., et al. CD44v/CD44s expression patterns are associated with the survival of pancreatic carcinoma patients. Diagn Pathol 9, 79, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tsuneki M., and Madri J.A. CD44 modulation of cell-cell and cell-matrix interactions, affecting survivin and hippo pathways. J Cell Physiol 231, 731, 2016 [DOI] [PubMed] [Google Scholar]
- 37. Moh M.C., and Shen S. The roles of cell adhesion molecules in tumor suppression and cell migration: a new paradox. Cell Adh Migr 3, 334, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Harari D., and Yarden Y. Molecular mechanisms underlying ErbB2/HER2 action in breast cancer. Oncogene 19, 6102, 2000 [DOI] [PubMed] [Google Scholar]
- 39. Wang K.H., Kao A.P., Chang C.C., et al. Increasing CD44+/CD24(−) tumor stem cells, and upregulation of COX-2 and HDAC6, as major functions of HER2 in breast tumorigenesis. Mol Cancer 9, 288, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Olsson E., Honeth G., Bendahl P.O., et al. CD44 isoforms are heterogeneously expressed in breast cancer and correlate with tumor subtypes and cancer stem cell markers. BMC Cancer 11, 418, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Williams K., Motiani K., Giridhar P.V., and Kasper S. CD44 integrates signaling in normal stem cell, cancer stem cell and (pre)metastatic niches. Exp Biol Med (Maywood) 238, 324, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]