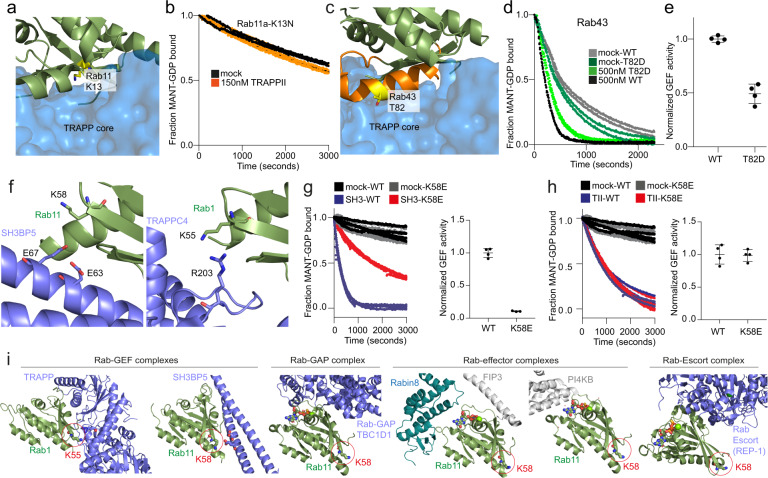
Fig. 5. Biochemical analysis of clinical Rab mutants shows specific altered GEF activity.
a Model of Rab11 bound to the TRAPP core, with the K13 residue that is mutated in developmental disorders (K13N). b In vitro GEF assay of TRAPPII WT (150 nM) for the clinically relevant mutation Rab11a K13N (4 μM) (n = 3). c Model of Rab43 bound to the TRAPP core, highlighting the residue T82 which can be phosphorylated by LRRK2. d Normalized GEF activity showing TRAPPII activation of WT Rab43 and Rab43-T82D phosphometic mutant (n = 2). e The rates of GEF exchange for WT Rab43 and phosphomimetic T82D normalized against WT. Error bars show SD (n = 4). f Model of SH3BP5 and the TRAPP core bound to Rab11 and Rab1, respectively (based on PDB models PDB:3CUE, 6DJL)38,52. The modified residue (K58) is labeled on both. g (left) Normalized GEF activity trace showing SH3BP5 (150 nM) activation of Rab11 WT (4 μM) and Rab11-K58E (4 μM) mutant (n = 3). (right) Rates of exchange were calculated and normalized against WT to determine the effect of the K58 mutant activation by SH3BP5. Error bars show SD (n = 4). h (left) Normalized GEF activity showing TRAPPII (150 nM) activation of Rab11 WT (4 μM) and Rab11-K58E (4 μM) mutant (n = 3). (right) Rates of exchange were calculated and normalized against WT to determine the effect of the K58 mutant activation by TRAPPII. Error bars show SD (n = 4). i Analysis of the interface of Rab11 with GEFs, GAPs, escort, and effector proteins compared to the location of the mutated K58 residue. The GEF complexes are from the structures of yeast TRAPP core bound to Rab1 (PDB: 3cue), and SH3BP5 bound to Rab11 (PDB: 6DJL)38,52. The GAP model was generated by aligning Rab11 with the Rab33 portion of the gyp1/Rab33 complex (PDB: 2G77)81. The structure of the TBC domain of the human TBC1D1 (PDB: 3QYE)82 was then superimposed on the Gyp1p TBC domain, and the cartoon TBC1D1 TBC domain is illustrated (blue). The effector models are from the structure of Rab11 bound to FIP3 and Rabin8, and Rab11 bound to PI4KB (PDB: 4UJ4, 4D0L)83,84. The escort complex was generated by superimposing the Rab7 component of a REP-1/Rab7 complex (PDB: 1VG0) onto Rab185.