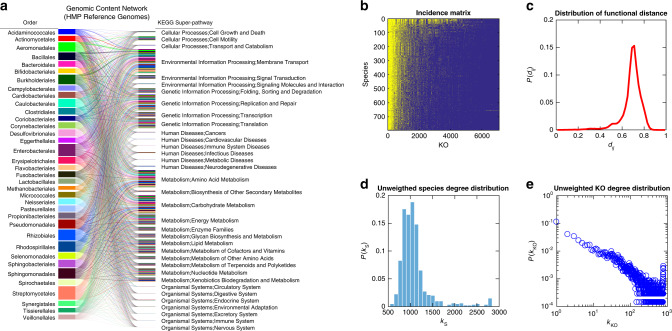
Fig. 2. The genomic content network (GCN) constructed from the Integrated Microbial Genomes and Microbiome (IMG/M) database has nested structure and heterogeneous gene degree distribution.
We use IMG/M-HMP, an IMG/M data mart that focuses on the Human Microbiome Project (HMP) generated metagenome data sets29 to construct the GCN. a For visualization purpose, we depict this reference GCN at the order level for taxon nodes and at the KEGG super-pathway level for function nodes. The bar height of each order corresponds to the average genome size of those species belonging to that order. The thickness of a link connecting an order and a KEGG super-pathway is proportional to the number of KOs that belong to that super-pathway, as well as the genomes of species in that order. The majority of the super-pathways shown here are related to the metabolic, environmental, and genetic processes performed by microbes. However, for a small number of taxa, as some of their genes have mammalian and/or human disease orthologs, we also identified several super-pathways involved in human diseases and higher-order organizational systems. See Supplementary Sec. 2.1 for the details of constructing this reference GCN. b The incidence matrix of this reference GCN is shown at the species-KO level, where the presence (or absence) of a link between a species and a KO is colored in yellow (or blue), respectively. We organized this matrix using the Nestedness Temperature Calculator to emphasize its nested structure31. The nestedness value (∼0.34712) of this network is calculated based on the classical NODF measure32 (see Methods for details). c The probability distribution of functional distances (dij) among different species. The bin size is 0.02. d The unweighted species degree distribution. Here, the unweighted degree of a species is the number of distinct KOs in its genome. e The unweighted KO-degree distribution. Here, the unweighted degree of a KO is the number of species whose genomes contain this KO.