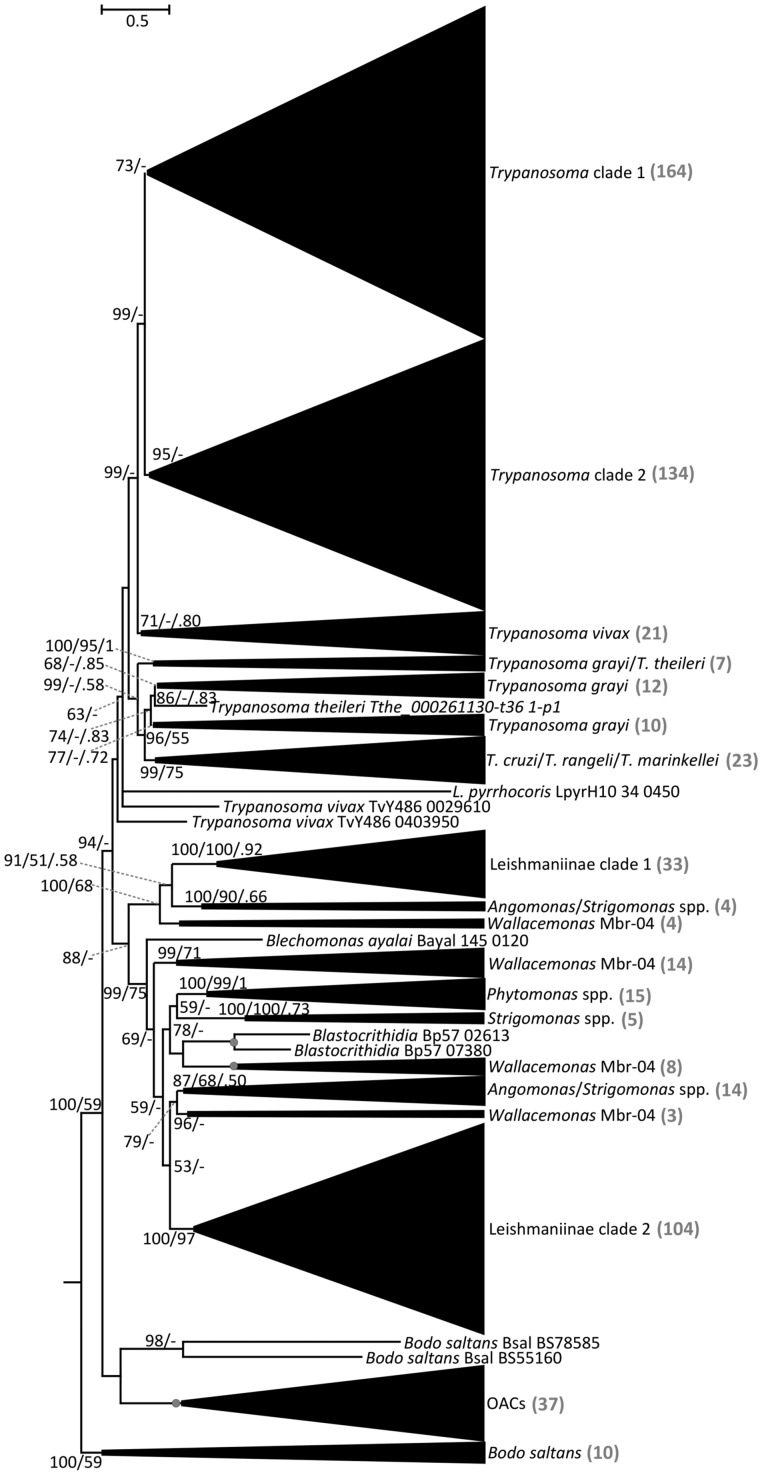
Fig. 1.
Phylogenetic tree of kinetoplastid ACs. Maximum-likelihood phylogenetic tree of kinetoplastid ACs based on a trimmed alignment of 631 sequences. Scale bar on top left represents 0.5 substitutions per site. The support values are in the following format: SH-aLRT support (%)/bootstrap support (%)/posterior probability. Only bootstrap support values ≥50% and posterior probabilities ≥0.50 are shown; nodes displaying maximal supports are marked with gray circles. “Trypanosoma clade 1” and “Trypanosoma clade 2” contain the genes of salivarian clades (T. congolense, T. brucei brucei, T. brucei gambiense, T. evansi, and T. equiperdum) upregulated in bloodstream and procyclic forms, respectively (see supplementary file S1, Supplementary Material online for details). OACs, oxygen-sensing adenylate cyclases from Leishmaniinae and other species (complete tree in fig 4). “Leishmaniinae clade 1” and “Leishmaniinae clade 2” contain homologs of leishmanial RAC-A and RAC-B sequences. The number of sequences within each collapsed clade is shown in gray color in the brackets.