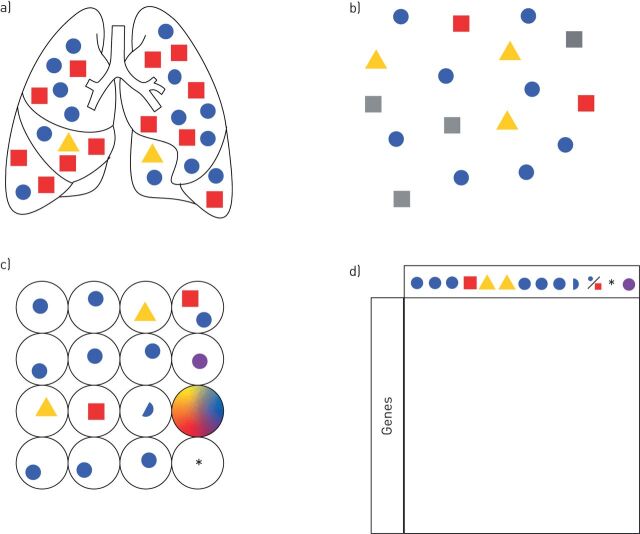
FIGURE 2.
Typical workflow of a single-cell RNA-sequencing experiment and possible pitfalls. a) Whole-lung tissue with highly abundant cellular components (blue circle, red square) and rare cell type (yellow triangle). b) A single-cell suspension is created through the mechanical and enzymatic disaggregation of the lung. Cell types that are fragile or difficult to liberate intact from the tissue (grey squares) may be underrepresented in the final dataset. In the lung, these cell types typically include alveolar type 1 epithelial cells and mesenchymal cells including fibroblasts. c) Individual cells are isolated into vessels (plate wells or droplets) with barcoded RNA primers and the cells are lysed to create a unique complementary DNA (cDNA) library for each cell. A number of errors can be introduced at this step, including the inclusion of two or more cells in a vessel, the inclusion of a cell fragment, the creation of a library from an empty vessel (*) or one containing ambient RNA (droplet with all colours), the inclusion of apoptotic cells or the induction of transcriptional changes as a result of processing (purple circle). d) cDNA libraries are sequenced and separated by barcode to get all the individual sequences for the individual cell. Sequences are aligned to a reference genome and successful alignments with known gene sequences are counted. This expression matrix is filtered by several quality criteria and used for downstream analyses.