Figure 2.
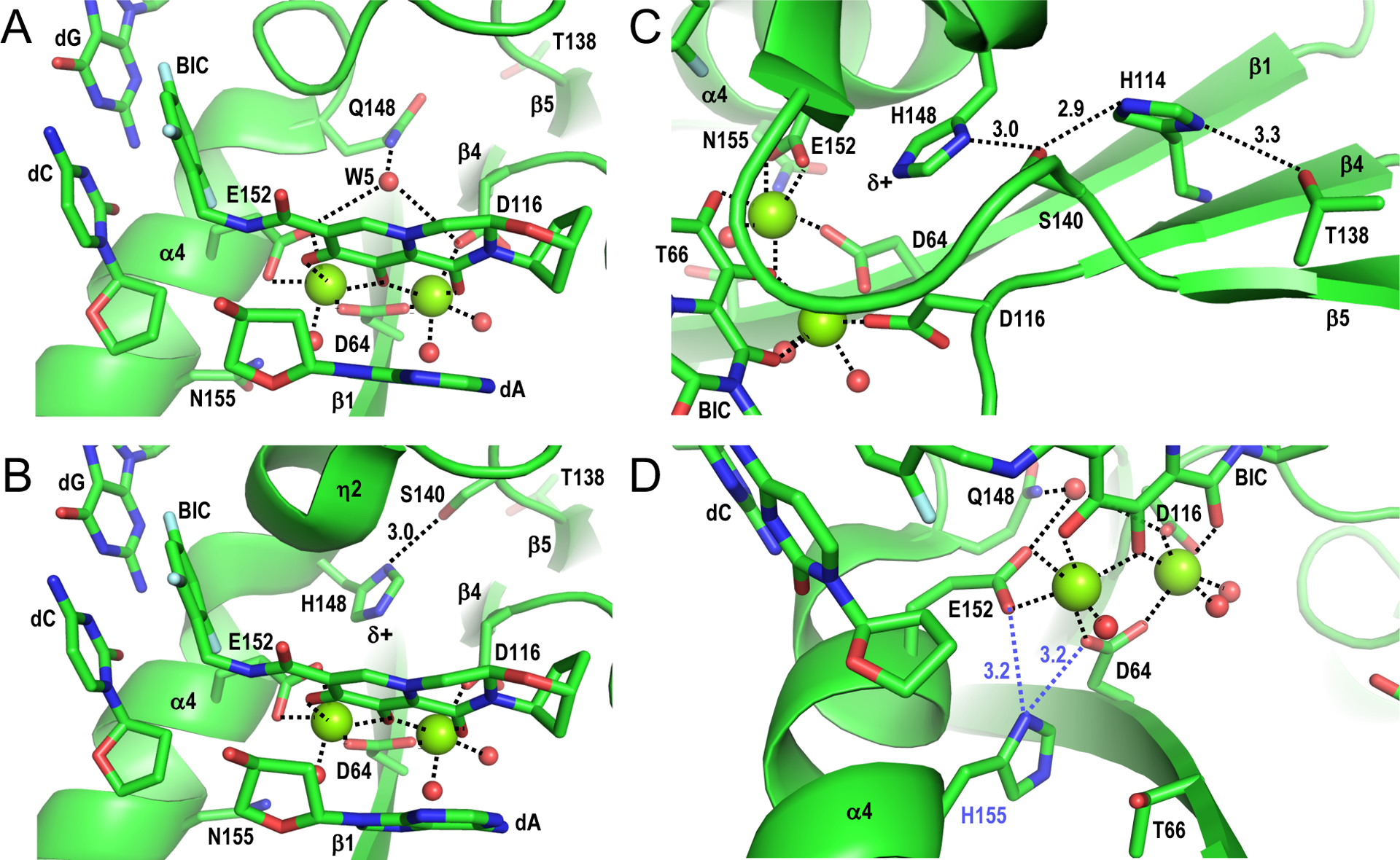
Structural basis of INSTI drug resistance. (A) Structure of SIVrcm intasome in complex with BIC (PDB code 6RWM) oriented to highlight magnesium ion interactions at the IN active site (dashed lines). Red spheres, water molecules; other labeling is the same as in Figure 1C. (B) Structure of the SIVrcm Q148H/G140S intasome bound to BIC (PDB code 6RWO; active site resolution 2.8 Å), oriented as in panel A. The distance between Ser140 Oγ and His148 Nδ1 is given in Å. (C) Same as in panel B, oriented to highlight the hydrogen bonding network involving IN residues Thr138, His114, Ser140, and His148. (D) Hypothetical interaction between mutant His155 side chain with SIVrcm IN active site. Asn155 within the SIVrcm intasome-BIC complex (PDB code 6RWM) was altered to His using the Wizard option of PyMOL [40], selecting the rotamer that most closely mimicked the rotameric configuration of Asn155. In this configuration, His155 (blue label) could potentially disrupt metal ion cluster stabilization by interacting with the carboxylate sidechain of active site residue Asp64 or Glu152 (dashed blue lines). Panel A-D structures were generated using PyMOL [40].
