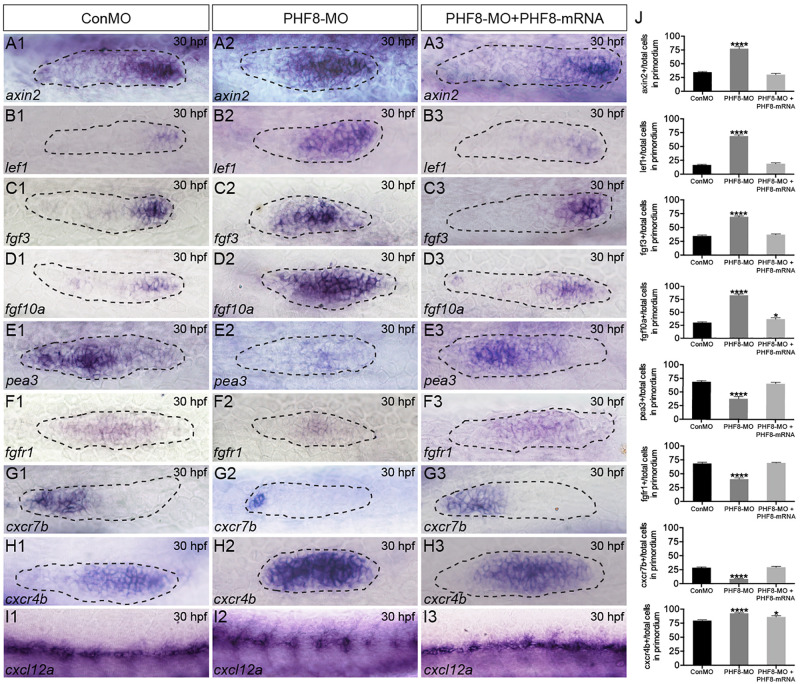
FIGURE 5.
Analysis of expression of genes of Wnt and FGF signaling pathways, as well as cxcl12/cxc7b/cxcr4b. (A–I) WISH was utilized to examine the expression of axin2 (A), lef1 (B), fgf3 (C), fgf10a (D), pea3 (E), fgfr1 (F), cxcr7b (G), cxcr4b (H), and cxcl12 (I) at 30 hpf. (J) The ratio of axin2, lef1, fgf3, fgf10a, pea3, fgfr1, cxcr7b, and cxcr4b-positive cells to total cells in the primordia of controls (ConMO; n = 10 embryos for axin2, 10 embryos for lef1, 12 embryos for fgf3, 12 embryos for fgf10a, 11 embryos for pea3, 9 embryos for fgfr1, 10 embryos for cxcr7b, 12 embryos for cxcr4b), PHF8 morphants (PHF8-MO; n = 10 embryos for axin2, 10 embryos for lef1, 14 embryos for fgf3, 17 embryos for fgf10a, 9 embryos for pea3, 9 embryos for fgfr1, 15 embryos for cxcr7b, 17 embryos for cxcr4b), and PHF8-MO co-injected with PHF8-mRNA embryo (PHF8-MO + PHF8-mRNA; n = 10 embryos for axin2, 10 embryos for lef1, 10 embryos for fgf3, 9 embryos for fgf10a, 9 embryos for pea3, 10 embryos for fgfr1, 12 embryos for cxcr7b, 12 embryos for cxcr4b). Data are expressed as mean ± SEM. *p < 0.05, ****p < 0.0001. All optical appearances elucidate lateral visions; anterior is on the left. The primordium is delineated with a dashed line.