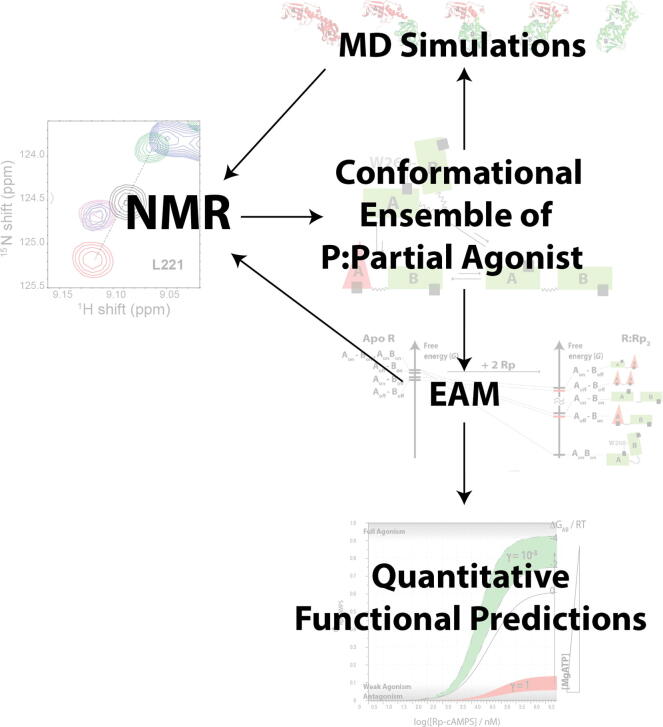
Fig. 11.
Synergies between NMR, MD and EAM enable quantitative modeling of enzyme function. NMR provides an initial map of the states within the conformational ensemble of a protein:partial-agonist complex, which serves as a basis to build initial structures for MD simulations and an EAM. The MD simulations serve as an effective means to generate refined targeted hypotheses to be tested by NMR. The EAM input parameters can often be measured by NMR. The fully parameterized EAM model enables bridging from protein dynamics to quantitative predictions of enzymatic function (e.g. kinase activity). Figures are adapted from Byun JA, Akimoto M, VanSchouwen B, Lazarou TS, Taylor SS, Melacini G. Allosteric pluripotency as revealed by protein kinase A. Sci Adv 2020;6:eabb1250. Reprinted with permission from AAAS.