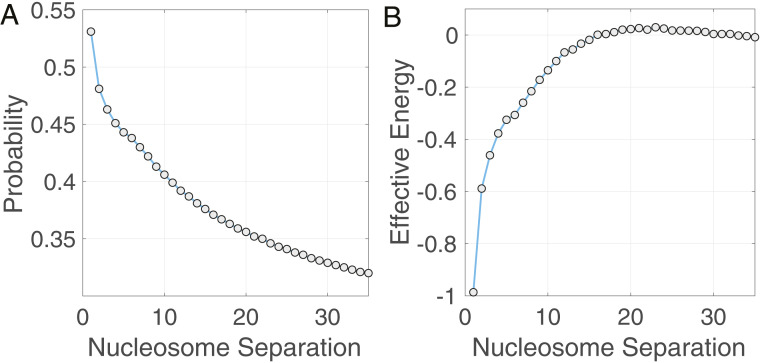
Fig. 6.
Analysis of H3K27me3 ChIP-seq data supports interaction between nonadjacent nucleosomes. (A) Probability for the H3K27me3 mark to simultaneously occur at two nucleosomes as a function of their separation length. (B) Effective interaction energy that measures the direct correlation between H3K27me3 occupancy at two nucleosomes as a function of their separation length. The energy profile supports the chromatin looping model. In contrast, for the linear model, only the energy between nearest-neighbor nucleosomes (a nucleosome separation of 1) has a nonzero value.