Fig. 5.
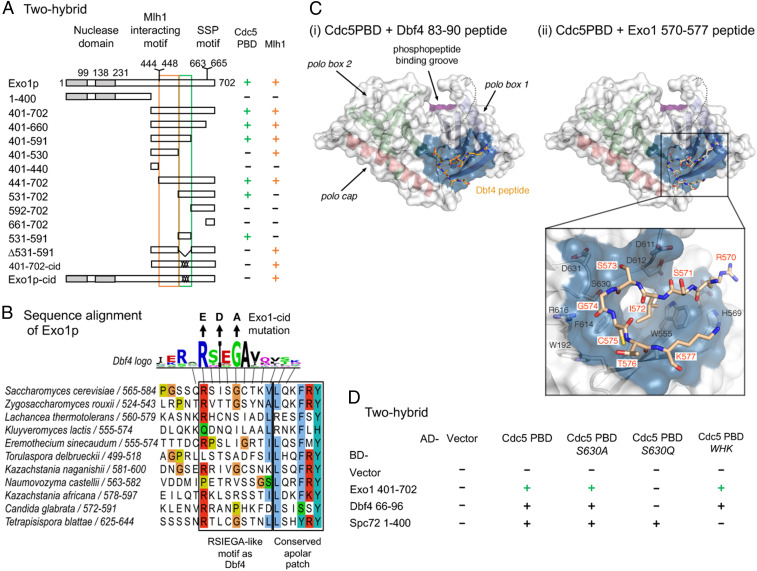
Cdc5 directly interacts with Exo1 through a noncanonical site. (A) Delineation of the Exo1 motif responsible for interaction with Cdc5 PBD by two-hybrid assays. The GAL4-BD fusions with indicated Exo1 fragments were tested in combination with a GAL4–AD–Cdc5–PBD fusion; “+” indicates an interaction. Exo1-cid: Cdc5 interaction-deficient. (B) Conservation of the Exo1 region interacting with Cdc5 PBD and illustration of the Exo1-cid mutation. The Dbf4 motif interacting with Cdc5 PBD (47) is indicated (consensus from 17 Saccharomycetaceae family species; see Materials and Methods). (C) Modeling of Dbf4 and Exo1 motifs on the crystal structure of Cdc5 PBD. (i) Crystal structure of the Cdc5 PBD bound to a Dbf4-derived peptide encompassing the RSIEGA motif (PDB ID code 6MF6) (48). The structural elements of the polo box domain are color coded. The region of the domain where phosphorylated substrates bind is labeled for reference. (ii) Model of the Cdc5–Exo1 interaction based on the crystal structure of the Cdc5–Dbf4 complex with Cdc5 shown in the same orientation and color scheme as in (i). The inset shows the residues mediating the interaction between the RSIEGA motif of Exo1 (red labels) and Cdc5 (black labels). (D) The same surface of Cdc5 used for interaction with Dbf4 is used for interaction with Exo1. The GAL4–BD fusions with indicated fragments were tested in combination with a GAL4–AD–Cdc5–PBD fusion with the indicated mutations (WHK stands for W517F H641A K643M); “+” indicates an interaction. See also SI Appendix, Figs. S7 and S8.