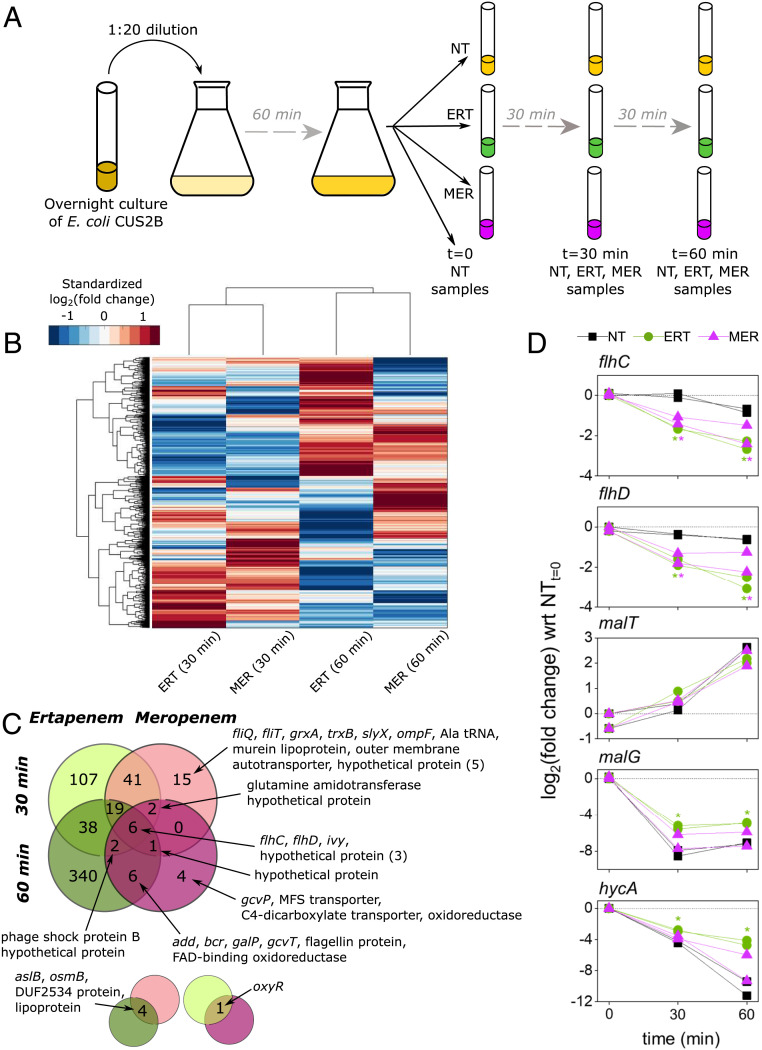
Fig. 2.
Total RNA sequencing uncovers ertapenem (ERT) response and identifies motility genes as resistance factors. (A) Overview of the sample collection protocol for RNA sequencing. Exponential-phase cells were grown with no antibiotic (NT), meropenem (MER), or ERT and collected after 0 (NT only), 30, and 60 min of growth in CAMHB. Two biological replicates were sampled for each condition. (B) Hierarchical clustering of gene expression values in ERT- and MER-treated E. coli CUS2B. Values are log2(fold change), with respect to the corresponding no treatment duplicates at the same time point. For visual clarity, the heat map has been standardized such that the mean is zero and the SD is one across each gene (each row). (C) The Venn diagram shows the degree to which significantly DE genes (expression levels compared with the no treatment condition at the given time point) overlapped across the four different conditions. Smaller pop outs are shown below to indicate overlaps on the main diagram diagonals. tRNA: transfer RNA; MFS: major facilitator superfamily; FAD: flavin adenine dinucleotide. (D) Time course of gene expression changes for the five transcripts that were explored further using PNA experiments. All conditions are normalized to the 0-min time point expression levels for the respective replicate, measured in duplicate just before antibiotic treatments were introduced. Asterisks indicate significant DE (q < 0.05) vs. the no treatment condition at the corresponding time point.