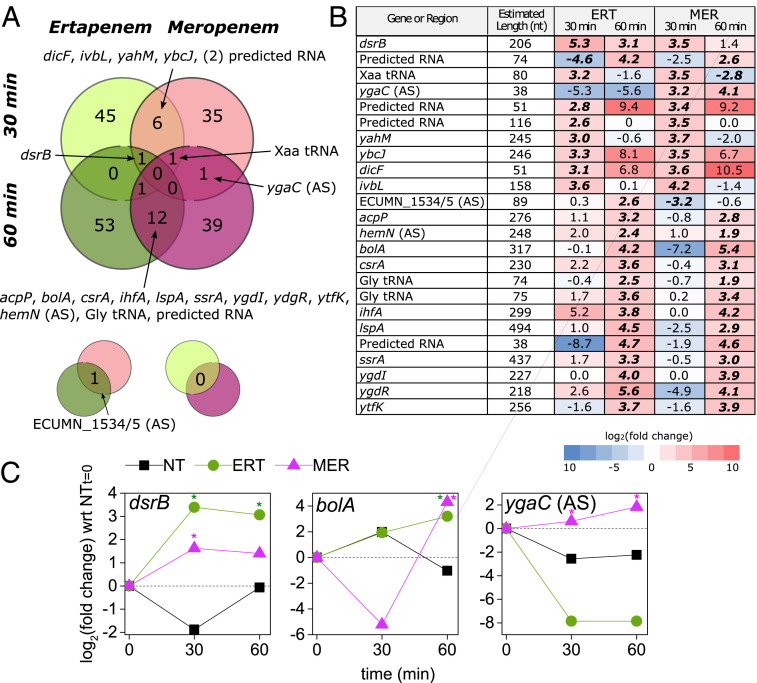
Fig. 3.
Short RNA sequencing identifies regulator genes and targets for PNA antibiotics. (A) Experimental setup was consistent between total RNA and short RNA sampling (Fig. 2A). Two biological replicates were sampled for each condition. The Venn diagram shows the degree to which significantly DE genes (expression levels compared with the no treatment [NT] condition at the given time point) overlapped across the four different conditions. Smaller pop outs are shown below to indicate overlaps on the main diagram diagonals. tRNA: transfer RNA. (B) Detail on the 22 RNAs that were DE in at least two conditions; log2(fold change) values here are with respect to the NT condition from the same time point. Bold italicized text indicates significant DE (q < 0.05) vs. the NT condition at the corresponding time point. (C) Time course of gene expression for three short RNA transcripts of interest. All conditions are normalized to the 0-min time point expression levels, measured in duplicate just before antibiotic treatments were introduced. ERT, ertapenem; MER, meropenem. *Significant DE (q < 0.05).