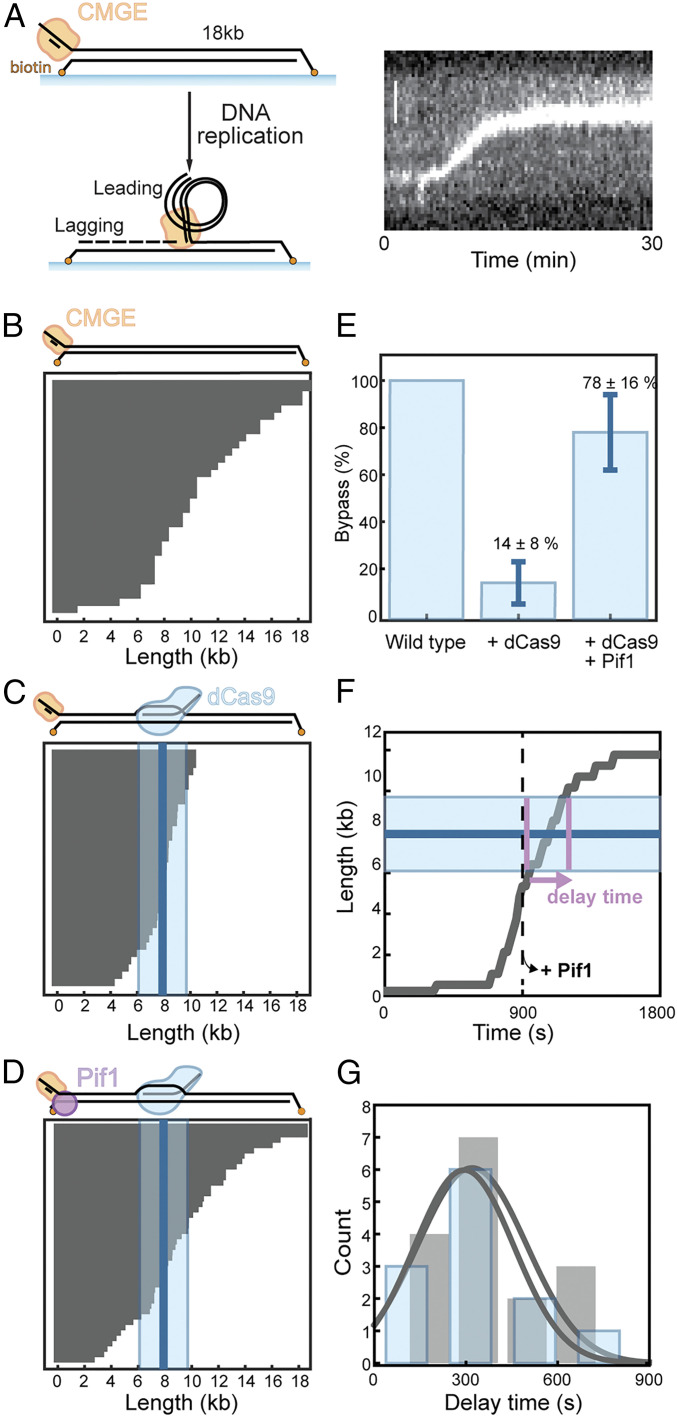
Fig. 4.
Single-molecule examination of Pif1-dependent replisome bypass of a dCas9 blocked site. (A) Schematic representation of the single-molecule assay (described in text) and a representative kymograph. Leading-strand DNA (stained with SYTOX Orange) accumulates along with a progressing replication fork. (B) Replication product lengths for replication without a dCas9 block. Every gray line represents the length of a single molecule. (C) Replication product lengths for replication in the presence of the dCas9 block bound to the leading strand. The blue line represents the average position of the dCas9 block. The SD is indicated by the light blue area. (D) Replication product lengths for replication in the presence of the dCas9 block where Pif1 was added after 15 min. (E) Efficiency of bypass of the dCas9 block for the experiments in B–D normalized to the experiment without block. We define a block as bypassed when a replisome moves past the average position of the block + SD (9.8 kb). For the reaction without dCas9 block (A), we define the “bypass” efficiency as the number of replisomes that move past 9.8 kb. The error bars represent the SEM from three experiments. (F) Representative single-molecule trajectory showing the position of the replisome as a function of time (gray line). The blue line represents the position of the dCas9 block. The SD is indicated by the light blue area. Pif1 was added after 15 min (indicated by the dashed line). We define the delay time as the time it takes the replisome to move through the area indicated by the SD of the block position.(indicated by the pink arrow). (G) Histograms of the delay time for replication without dCas9 block (gray) and replication in which a leading strand block is resolved by Pif1 (blue). Again, for the reaction without dCas9 block, we define “bypass” when replisomes move past 9.8 kb. Gaussian fits (represented by the gray lines) give average delay times of 294 ± 137 s (mean ± SEM) for replication without block and 320 ± 150 s (mean ± SEM) for blocked replication resolved by Pif1.