Significance
Identifying signaling pathways contributing to resistance to anti-TNF therapy in rheumatoid arthritis is crucial for the development of new therapeutic strategies for refractory rheumatoid arthritis. Th17 cells, a subset of proinflammatory CD4+ T cells, are implicated in the pathogenesis of the disease. We analyzed the gene expression profiles of Th17-enriched CD4+ T cells in anti-TNF–treated patients with rheumatoid arthritis and found that the elevated expression levels of transcription factor USF2 in anti-TNF refractory patients were associated with increased proinflammatory signaling of Th17 cells. USF2-knockdown experiments in Th17 cells revealed that USF2 promotes the pathogenicity of Th17 cells. These findings have implications for the development of new therapeutic strategies for refractory rheumatoid arthritis.
Keywords: USF2, Th17, rheumatoid arthritis, proinflammatory, gene expression
Abstract
IL-17–producing Th17 cells are implicated in the pathogenesis of rheumatoid arthritis (RA) and TNF-α, a proinflammatory cytokine in the rheumatoid joint, facilitates Th17 differentiation. Anti-TNF therapy ameliorates disease in many patients with rheumatoid arthritis (RA). However, a significant proportion of patients do not respond to this therapy. The impact of anti-TNF therapy on Th17 responses in RA is not well understood. We conducted high-throughput gene expression analysis of Th17-enriched CCR6+CXCR3−CD45RA− CD4+ T (CCR6+ T) cells isolated from anti-TNF–treated RA patients classified as responders or nonresponders to therapy. CCR6+ T cells from responders and nonresponders had distinct gene expression profiles. Proinflammatory signaling was elevated in the CCR6+ T cells of nonresponders, and pathogenic Th17 signature genes were up-regulated in these cells. Gene set enrichment analysis on these signature genes identified transcription factor USF2 as their upstream regulator, which was also increased in nonresponders. Importantly, short hairpin RNA targeting USF2 in pathogenic Th17 cells led to reduced expression of proinflammatory cytokines IL-17A, IFN-γ, IL-22, and granulocyte-macrophage colony-stimulating factor (GM-CSF) as well as transcription factor T-bet. Together, our results revealed inadequate suppression of Th17 responses by anti-TNF in nonresponders, and direct targeting of the USF2-signaling pathway may be a potential therapeutic approach in the anti-TNF refractory RA.
Rheumatoid arthritis (RA) is a chronic autoimmune disorder affecting joints (1). Cytokines are involved in the pathogenesis of RA, including tumor necrosis factor-α (TNF-α), IL-1, IL-6, IL-17, IL-23, and granulocyte-macrophage colony-stimulating factor (GM-CSF) (2–4). TNF-α is primarily produced by activated macrophages and monocytes, although it may also be produced by lymphocytes and other cell types including T helper 17 cells (Th17) (3, 5–7), an IL-17–producing CD4+ T cell subset which was first reported in murine autoimmune models in 2005 (9, 10). As one of the major proinflammatory cytokines present in the rheumatoid joint, TNF-α has proven to be a good therapeutic target for RA therapy, and TNF-α inhibitors effectively block disease progression and improve physical function (8). Th17 cells have been linked to autoimmune diseases including RA, multiple sclerosis, systemic lupus erythematosus, psoriasis, inflammatory bowel disease, and Crohn’s disease (11, 12). Increased frequency of Th17 cells and elevated IL-17 levels have been found in the peripheral blood of RA patients (13, 14). The increased frequency of Th17 cells correlates with the number of swollen joints and serum levels of C-reactive protein (15) and IL-17. Th17 cells in inflamed joints in RA orchestrates the chronic inflammation by stimulating fibroblast-like synoviocytes to produce GM-CSF and expand proinflammatory-secreting synovial-resident innate lymphoid cells (16). When first discovered, Th17 cells were considered a homogenous proinflammatory population (9, 10). Shortly thereafter, an antiinflammatory subset of Th17 cells that coproduced IL-10 was identified (17), while proinflammatory/pathogenic Th17 cells are shown to express higher levels of IFN-γ (17–19). Therapeutic studies also reveal the complexity of Th17 function. Anti–IL-17 therapy ameliorates psoriasis, but blocking the IL-17–signaling pathway in Crohn’s disease is either ineffective or exacerbates diseases (20–22). Thus, it is not only important to determine Th17 cell frequency and IL-17 levels in patients with RA, but also to evaluate the proinflammatory capacity of Th17 cells (18, 19). In juvenile idiopathic arthritis, IFN-γ–secreting Th17 cells are highly enriched in the synovial fluid (23), and IFN-γ–negative Th17 cells can be converted to IFN-γ–secreting Th17 cells under conditions of the disease flare (24). In adult RA patients, studies indicate the migration of IFN-γ–secreting Th17 cells to synovial tissue of inflammation (25). These finding suggest the association of proinflammatory Th17 cells in RA pathogenesis and inhibition of their function is a therapeutic approach worthy of exploration.
Upon T cell receptor activation, IL-6 and TGF-β1 induce nonpathogenic Th17 differentiation, while the cytokine combination of IL-1β, IL-6, and IL-23 leads to pathogenic Th17 differentiation (18, 26). TNF-α promotes Th17 differentiation in RA via inducing stromal cells and monocytes to secrete proinflammatory cytokines IL-1β, IL-6, etc. (27, 28). A recent clinical study has reported that TNF-α inhibitor infliximab reduces the frequency of peripheral Th17 cells and IL-17 level in patients with RA (15), which implies a direct or indirect upstream therapeutic effect of anti–TNF-α on Th17 differentiation. In other diseases, TNF-α and IL-17 have been shown to have synergistic effects to amplify proinflammatory signals. In psoriasis both TNF-α and IL-17 are overexpressed in the skin lesions, and they act synergistically to affect cytokine production (29). An in vitro study of intervertebral disk cells also demonstrated that TNF-α and IL-17 synergistically facilitate inflammatory mediator release (30). Gene expression profiling of CCR6+CXCR3−CD4+ T cells from anti–TNF-α–treated RA patients, which are enriched for Th17 cells, found that these treated patients still display enhanced gene expression of IL17, RORC, IL22, and IL23R compared to healthy controls (31). A proof-of-concept clinical study has shown that the combination of anti–IL-17 and anti-TNF treatment effectively reduces disease activity in RA patients with inadequate response to anti-TNF treatment alone (32). Taken together, these studies indicate that the TNF-α and IL-17/Th17 pathways do not completely converge in RA pathogenesis and that the IL-17/Th17 pathway is a nonredundant therapeutic target for the disease.
A lack of response to TNF-α inhibitors in a significant portion of patients is a serious problem in clinical rheumatology (33, 34). Investigation of responders and nonresponders to anti-TNF therapy can help to identify the pathways and key regulatory genes involved in refractoriness to the treatment. Here, we study the gene expression of Th17-enriched CCR6+CXCR3− memory CD4+ T (CCR6+ T) cells from responders and nonresponders to anti-TNF therapy in RA patients as well as Th17 cells induced in vitro. We identified the transcription factor USF2 to fuel-activated proinflammatory signaling pathways in anti-TNF refractory patients and experimentally confirmed the role of USF2 to control the expression of proinflammatory cytokines in pathogenic Th17 cells.
Results
Enrichment of Th17 Subsets in CD4+CCR6+CXCR3− T Cells.
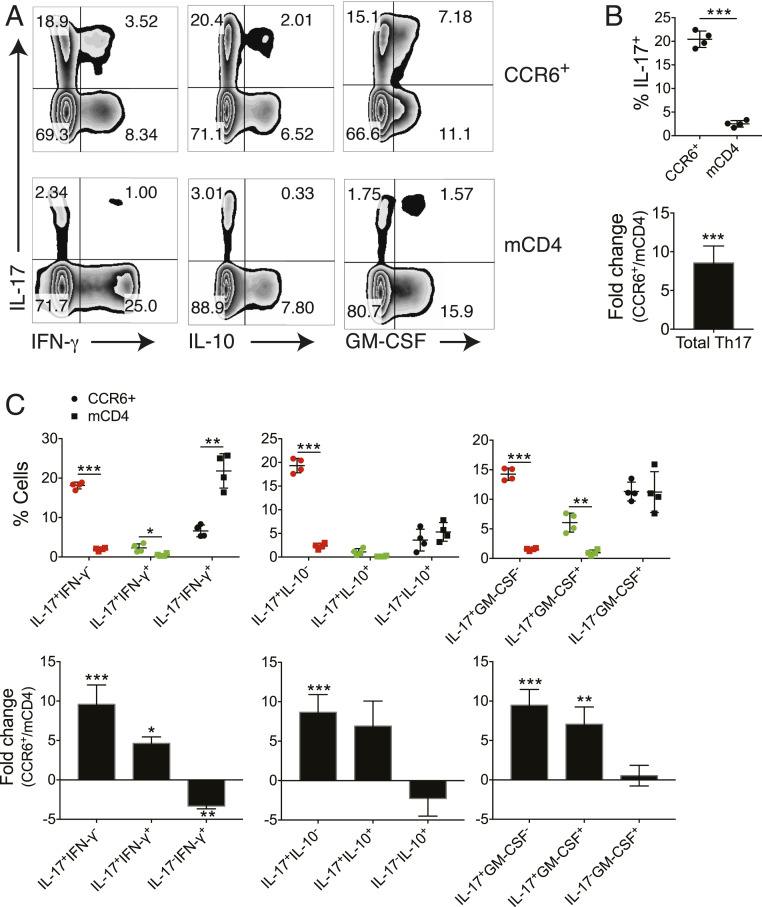
The expression of the CC chemokine receptor CCR6 is strongly correlated with both mouse and human Th17 cells (35, 36) and thus has been used as a cell surface marker to enrich Th17 cells given that there are no unique cell surface markers available for the Th17 subset. Based on the differential expression of chemokine receptors and other cell surface antigens, we utilized antibody-conjugated magnetic beads by negative selection to deplete non-CD4+ T cells by negative selection and to isolate Th17-enriched CCR6+CXCR3− memory CD4+ T cells by positive selection (see Materials and Methods) (SI Appendix, Fig. S1 A and B). Cells isolated in this manner are only bound by anti-CCR6 antibody to minimize unwanted manipulation and are hereafter referred to as CCR6+ T cells. To assess the efficiency of this approach for Th17 enrichment, we isolated total memory CD4+ T (mCD4) cells and CCR6+ T cells from the blood of four healthy donors and stained cells for the intracellular production of IL-17, IFN-γ, IL-10, and GM-CSF (Fig. 1). Compared to mCD4 cells, the Th17 frequency in CCR6+ T cells was increased by 7- to 12-fold (Fig. 1 A and B), and Th1 (IL-17−IFN-γ+) frequency was reduced by ∼70% (Fig. 1 A and C). The Th17/Th1 ratio was increased from 1/8.7 in mCD4 cells to 3.1/1 in CCR6+ cells with a fold change of 26.7. It has been reported that CXCR3 is rapidly induced in naive T cells upon activation, and its expression remains high on IFN-γ–secreting Th1 and IFN-γ+ Th17 cells (26, 37). However, IFN-γ+ Th17 cells were still enriched in CXCR3− CCR6+ T cells (Fig. 1 A and C and SI Appendix, Fig. S1B), demonstrating that, like Th1 cells, not all IFN-γ+ Th17 cells express cell surface CXCR3. Analysis of IL-10 and GM-CSF secretion in Th17 cells showed that the IL-10−, IL-10+, GM-CSF−, and GM-CSF+ Th17 subsets were also enriched in CCR6+ T cells. Thus, the CCR6 selection and CXCR3 depletion effectively enriched various Th17 subsets from peripheral blood.
Fig. 1.
Enriched Th17 and reduced Th1 populations in CD4+CCR6+CXCR3− T cells. CCR6+CXCR3− memory CD4+ T (CCR6+) cells and mCD4 T cells were isolated from the peripheral blood of four healthy donors and stimulated with PMA and ionomycin for 4 h. Production of cytokines in CD4+ T cells were assessed by flow cytometry with intracellular cytokine staining assay. (A) IL-17, IFN-γ, IL-10, and GM-CSF production in CCR6+ and mCD4 cells. Dot plots shown were gated on CD4+ cells from one representative individual. (B) Frequency (Upper) and fold change of the frequency (CCR6+ vs. mCD4) (Lower) of total Th17 cells in CCR6+ and mCD4 cells. (C) Frequency (Upper) and fold change of the frequency (Lower) on various cytokine-secreting Th-subsets. *P < 0.05, **P < 0.005, ***P < 0.0005, two-tailed, paired Student’s t test on each cell subset between CCR6+ and mCD4 cells (individuals n = 4, mean ± SD).
Distinct Gene Expression in Responding and Nonresponding RA Patients to Anti-TNF Therapy.
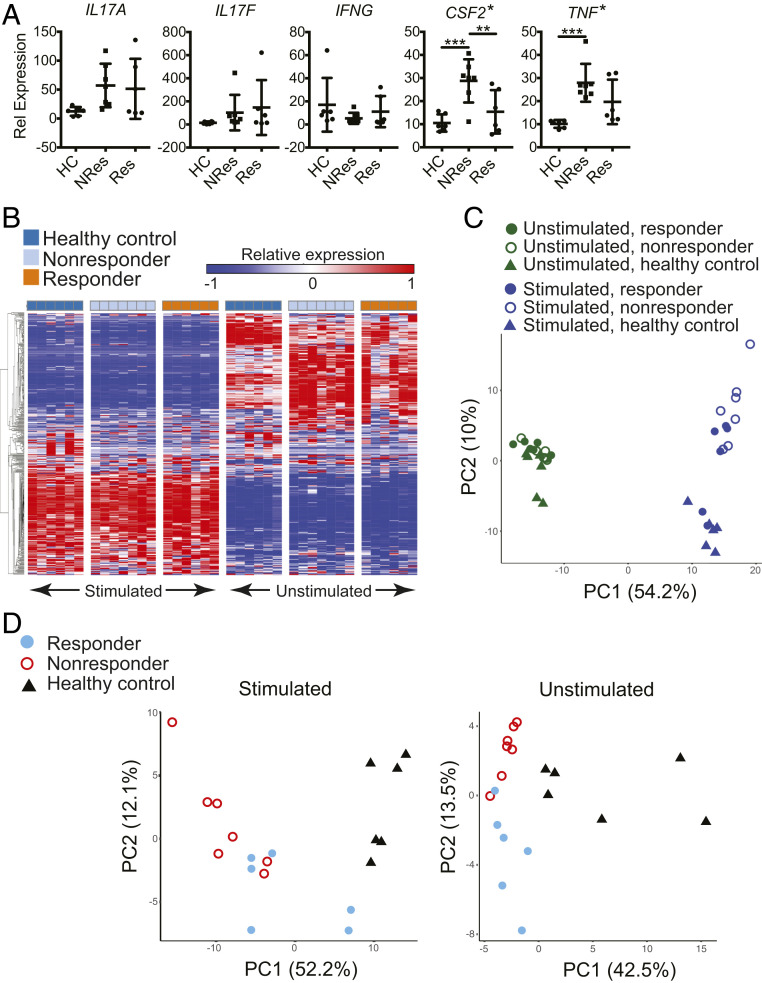
To assess the difference in gene expression of CCR6+ T cells between RA patients who responded or did not respond to anti-TNF treatment, we isolated CCR6+ T cells from six responding patients, seven nonresponding patients, and six healthy controls (Table 1). CCR6+ T cells were either activated with phorbol 12-myristate 13-acetate (PMA) / ionomycin for 4 h or left untreated and used as unstimulated controls. qPCR analysis of the activated cells showed that the expression levels of IL17A, IL17F, and IFNG were not different among these three groups. The CSF2 levels in both responding and nonresponding patients were elevated with nonresponders showing the trend of highest expression. The levels of TNF messenger RNA (mRNA) in the nonresponders but not in the responders were significantly higher compared to healthy controls (Fig. 2A). We also subjected the cells to the nCounter Gene Expression Analysis using the custom CodeSet HuTH17 that detects 418 genes associated with human T-helper (TH) cell differentiation and activation (19). For the 19 PMA/ionomycin stimulated and 19 unstimulated CCR6+ T cell samples we analyzed, 397 of 418 genes showed maximum expression levels across all samples above background and were selected for further analysis. The expression levels of IL17A, IL17F, IFNG, CSF2, and TNF in stimulated CCR6+ T cells were consistent with the results obtained with qPCR analysis (SI Appendix, Fig. S2). Supervised clustering (Fig. 2B) and principal component analysis (PCA) (Fig. 2C) of the 397-gene expression profiles of the 38 individual samples demonstrated distinct transcriptional characteristics between stimulated and unstimulated cells; in contrast, the difference among responding, nonresponding patients, and healthy controls was relatively minor. Nevertheless, the PCA plot clearly segregated the nonresponders from healthy controls in the stimulated samples. To better reveal the difference among sample groups, we analyzed stimulated and unstimulated samples independently. Here we focused on the genes with one-way ANOVA P value < 0.05 among the three sample groups of either stimulated or unstimulated cells. Based on this restriction, 172 and 74 genes were selected for the stimulated (SI Appendix, Table S1) and unstimulated samples (SI Appendix, Table S2), respectively. PCA showed that healthy controls were well separated from RA patients for both stimulated and unstimulated CCR6+ T cells, while responders were well separated from nonresponders for unstimulated cells and, to a lesser extent, for stimulated cells (Fig. 2D). These results demonstrated that differential gene expression in CCR6+ T cells not only distinguished patients from healthy donors but also responders from nonresponders to anti-TNF treatment.
Table 1.
Demographics of patients with rheumatoid arthritis and healthy controls
| RA patients | Healthy controls | ||
| Responders | Nonresponders | ||
| Participants, n | 6 | 7 | 6 |
| Gender F/M, n | 6/0 | 7/0 | 6/0 |
| Age, y | 63 ± 2 | 65 ± 14 | 57 ± 10 |
| Disease duration, y | 17 ± 8 | 28 ± 16 | n.a. |
| DAS28* | 2.0 ± 0.7 | 3.8 ± 0.6 | n.a. |
| RF-positive, n (%) | 3 (50%) | 6 (86%) | 0 (0%) |
| ACPA-positive, n | 4 (67%) | 4 (57%) | 0/6 |
| MTX, mg/wk† | 14 ± 5.18 | 16 ± 6.10 | n.a. |
| Infliximab total, mg | 8,367 ± 4,234 | 8,257 ± 7,051 | n.a. |
All RA patients received infliximab injection i.v. at a dose of 200 mg every 8 wk. DAS28, 28 joint disease activity score. RF, rheumatoid factor. ACPA, anticitrullinated protein antibodies. MTX, methotrexate. n.a., not applicable.
P = 0.0005 between responders and nonresponders (Student’s unpaired, two-tailed, heteroscedastic test).
One patient was treated with mycophenolate mofetil instead of MTX.
Fig. 2.
Differential gene expression in CCR6+ cells distinguishes among responding and nonresponding RA patients and healthy donors. (A) qPCR analysis of CCR6+ cells isolated from healthy donors and patients responding and nonresponding to anti-TNF treatment. Cells were stimulated with PMA and ionomycin for 4 h. *, one-way ANOVA, P < 0.005; **, Tukey’s multiple comparison test, P < 0.05; and ***, Tukey’s multiple comparison test, P < 0.005 (mean ± SD). (B–D) Gene expression profiles of CCR6+ cells, with (stimulated) or without (unstimulated) PMA and ionomycin for 4 h, were obtained using the nCounter (nanoString Technologies) CodeSet HuTH17. The 397 out of the 418 measured genes in the CodeSet that showed maximum expression levels across all samples above background were selected for (B) supervised hierarchical clustering and (C) PCA of the individual samples. (D) Genes with one-way ANOVA P value < 0.05 among the three sample groups under either stimulated or unstimulated condition were selected for PCA, which resulted in 172 genes for stimulated cells and 74 for unstimulated cells. (Healthy controls, n = 6; nonresponders, n = 7; responders, n = 6).
Enhanced Proinflammatory Signaling in Nonresponding Patients.
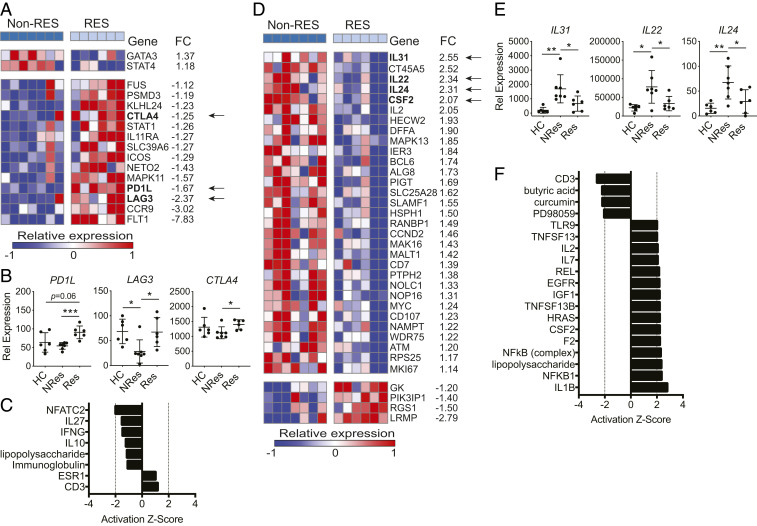
In unstimulated CCR6+ T cells, 16 of the 397 genes were differentially expressed between nonresponders vs. responders with 2 up-regulated and 14 down-regulated in the nonresponding patients. The PD1L, LAG3, and CTLA4 genes, which encode checkpoint inhibitors and are associated with T cell exhaustion (38, 39) and FoxP3+ CD4+ Treg-mediated suppression (40), were among the 14 down-regulated genes in nonresponding patients (Fig. 3A). Yet, PD1L, LAG3, and CTLA4 displayed different patterns of relative expression among healthy controls and RA patients (Fig. 3B). No significant difference of PD1L expression was observed between healthy controls and nonresponding patients, but its expression was up-regulated in responding patients. For LAG3, nonresponding patients displayed repressed expression compared to healthy controls and responding patients, while its expression was comparable between the two latter groups. The expression pattern of CTLA4 was more similar to that of PD1L than that of LAG3, with it not showing a reduced expression in nonresponding patients compared to healthy controls. The Ingenuity Pathway Analysis (IPA) upstream regulator analysis on the 16 DE genes identified the upstream regulator NFATC2 inhibited in nonresponders (Fig. 3C). NFATC2 is an intrinsic negative regulator of T cell activation (41) and facilitates Treg induction and function (42). The reduced expression of LAG3 and CTLA4 and inactivated NFATC2-signaling pathway in the CCR6+ T of nonresponders suggested a lower Treg activity compared to responders. In stimulated CCR6+ T cells, 35 genes were differentially expressed between nonresponders and responders (Fig. 3D). Among the top six genes up-regulated in nonresponders with a fold change above 2, four genes encode proinflammatory cytokines IL-31 (43), IL-22 (44), IL-24 (45), and GM-CSF; one encodes T cell growth factor IL-2. While the expression of IL31, IL22, IL24, and CFS2 was increased in nonresponding patients, their expression levels between responding patients and healthy controls were comparable (Figs. 2A and 3E). Among the DE genes, IL22, CSF2, IL2, and SLAMF1 are known pathogenic Th17 signature genes (19). The IPA upstream regulator analysis of the 35 DE genes showed that IL-1β and NF-κB were the top two activated upstream regulators; GM-CSF was also among the activated upstream regulators. In contrast, butyric acid (46, 47) and curcumin (48–50), which activate antiinflammatory pathways, were among the inhibited upstream regulators (Fig. 3F). These results showed that the anti-TNF treatment was inadequate to down-regulate the abnormally elevated immune responses in the CCR6+ T cells of nonresponding patients compared to that of responders.
Fig. 3.
Proinflammatory pathways are activated in CCR6+ cells from nonresponders. (A) Differentially expressed (DE) genes (nCounter) in unstimulated CCR6+ cells from nonresponder vs. responder comparison (Non-Res, nonresponder; Res, responder; FC, fold change; unpaired two-tailed Student’s t test P < 0.05). (B) Expression of PD1L, LAG3, and CTLA4 in unstimulated CCR6+ cells (nCounter). Unpaired two-tailed Student’s t test, *P < 0.05 and ***P < 0.0005. (C) IPA upstream regulator analysis on DE genes shown in A. (D) DE genes (nCounter) in stimulated CCR6+ cells from nonresponder vs. responder comparison (P < 0.05). (E) Expression of IL31, IL22, and IL24 in stimulated CCR6+ cells (nCounter). (F) IPA upstream regulator analysis on DE genes shown in C. Unpaired two-tailed Student’s t test, *P < 0.05 and **P < 0.005.
Enriched Pathogenic Th17 Gene Signature in Nonresponders.
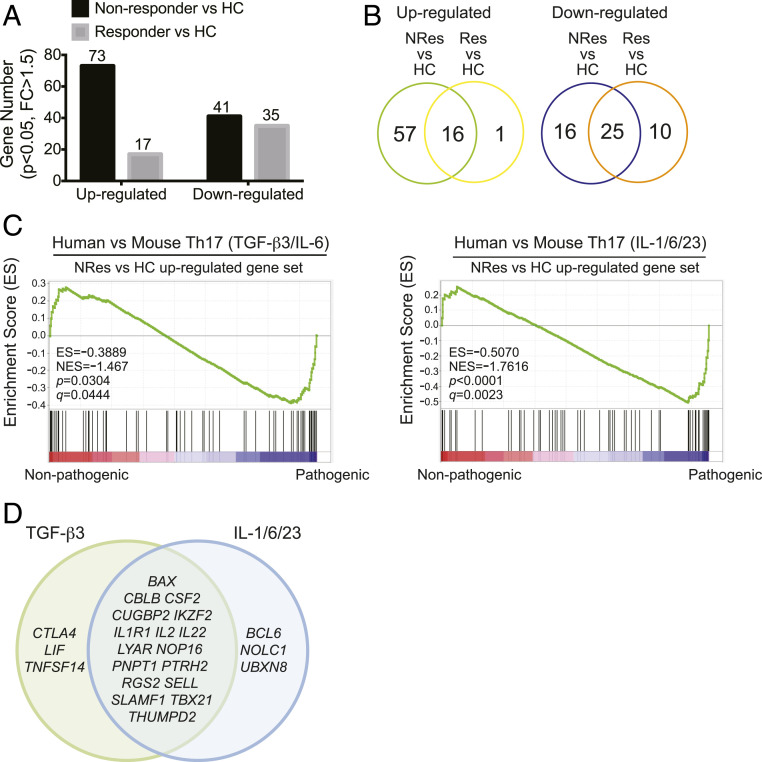
To interrogate whether the enhanced proinflammatory activity in the CCR6+ T cells of nonresponders was related to the pathogenicity of Th17 cells, we performed gene set enrichment analysis (GSEA) to cross-examine the similarities in signature genes between mouse pathogenic Th17 cells and the CCR6+ T cells of responders and nonresponders. Since many of the Th17-associated genes were primarily expressed in stimulated cells (SI Appendix, Fig. S3), we performed the GSEA with the gene expression profiles of stimulated CCR6+ T cells. First, we identified the DE genes between nonresponders vs. healthy controls and between responders vs. healthy controls. Compared to healthy controls, 73 genes were up-regulated and 41 down-regulated in nonresponding patients, whereas only 17 genes were up-regulated and 35 down-regulated in responding patients (Fig. 4A). These four gene sets formed the up-regulated and down-regulated gene signatures of nonresponders and responders, respectively. The up-regulated gene signature of responders was smaller than and largely included in that of nonresponders, with 16 out of the 17 signature genes present in the up-regulated gene signature of nonresponders, while the overlap in the down-regulated gene signatures was less (Fig. 4B and SI Appendix, Tables S3 and S4). Treating murine naive CD4+ T cells with TGF-β3/IL-6 or IL-1/IL-6/IL-23 cytokine mixtures induces pathogenic Th17 differentiation, whereas TGF-β1/IL-6 induces nonpathogenic Th17 cells, and the whole-genome microarray data on these murine Th17 populations are publicly available (18). Thus, the four gene signatures of nonresponders and responders were analyzed for gene set enrichment by being compared to murine TGF-β3/IL-6–induced Th17 cells (pathogenic) vs. TGF-β1/IL-6–induced Th17 cells (nonpathogenic) (scenario 1); and IL-1/IL-6/IL-23–induced Th17 cells (pathogenic) vs. TGF-β1/IL-6–induced Th17 (nonpathogenic) (scenario 2). GSEA results demonstrated that genes up-regulated in nonresponding patients were enriched in the mouse pathogenic vs. nonpathogenic Th17 comparison in both scenarios, but genes up-regulated in responding patients were not enriched (Fig. 4C). The enriched leading-edge genes of nonresponding patients identified by the two comparative scenarios were largely overlapping (Fig. 4D). Genes down-regulated in nonresponding patients or in responding patients were both enriched in the mouse pathogenic vs. nonpathogenic Th17 cell comparison (SI Appendix, Fig. S4A), and the enriched genes were largely overlapping between the nonresponder vs. mouse Th17 and responder vs. mouse Th17 comparisons for both scenarios (SI Appendix, Fig. S4B). We conducted these analyses to probe the key genes and pathways that were potentially involved in resistance to anti-TNF treatment in RA. These results demonstrated that the gene expression disparity in CCR6+ T cells between anti-TNF responding and nonresponding patients relative to pathogenic Th17 cells was found in up-regulated but not down-regulated signature genes, and the up-regulated leading-edge genes of CCR6+ T cells in nonresponders could be utilized to identify the plausible pathways that modulated the pathogenicity of Th17 cells to help establish the resistance to anti-TNF therapy in RA.
Fig. 4.
The up-regulated gene signature of CCR6+ cells in nonresponders but not responders is enriched in pathogenic Th17 cells. (A) The numbers of up-regulated and down-regulated DE genes in stimulated CCR6+ cells from nonresponder vs. healthy control comparison and responder vs. healthy control comparison (unpaired two-tailed Student’s t test, P < 0.05, FC ≥ 1.5) (nCounter). (B) Venn Diagram representations of the numbers of overlapped DE genes from the comparisons in A. (C) GSEA: enrichment of the up-regulated gene set of nonresponder vs. HC comparison in TGF-β3/IL-6 (Left) or IL-1/IL-6/IL-23 (Right) induced mouse pathogenic Th17 cells. (D) Venn diagram representations of leading-edge genes (enriched signature genes) in C: TGF-β3, genes enriched in TGF-β3/IL-6–induced Th17 cells; IL-1/6/23, genes enriched in the IL-1/IL-6/IL-23–induced Th17 cells. (NRes, nonresponder; Res, responder; HC, healthy donor; NES, normalized enrichment score).
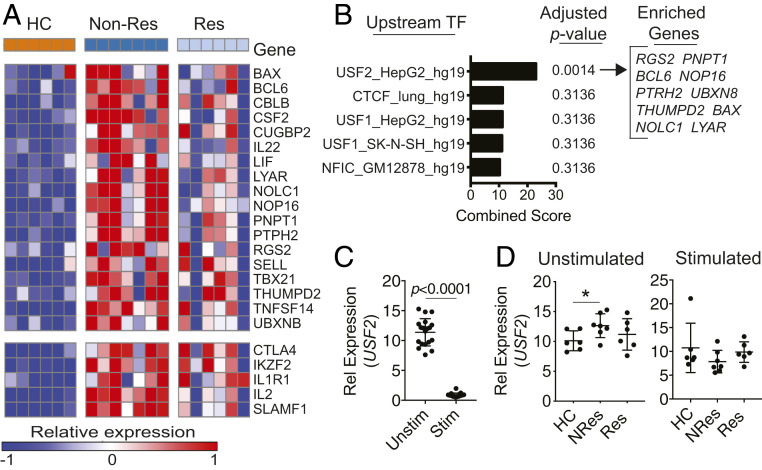
Identification of USF2 as the Upstream Regulator of the Refractory Gene Signature.
Merging the leading-edge genes listed in Fig. 4D, we obtained a molecular signature of CCR6+ T cells, which comprised 23-enriched up-regulated genes of nonresponding patients related to Th17 pathogenicity in contrast to responding patients. The expression of these up-regulated signature genes was progressively increased from healthy controls to responding and nonresponding patients (Fig. 5A). Comparing this gene signature to the DE genes between responders vs. healthy controls (SI Appendix, Table S4), we found five overlapping genes (CTLA4, IKZF2, IL1R1, IL2, and SLAMF1) (Fig. 5 A, Lower). To ensure that the signature genes we identified were unique for nonresponders, we removed the five overlapping genes and formed an anti-TNF refractory gene signature with the remaining 18 signature genes (Fig. 5 A, Upper). In the next step, we interrogated the obtained refractory gene signature for upstream transcription factors involved in nonresponsiveness to anti-TNF treatment with the Enrichr ENCODE TF CHIP-SEq 2015 analysis (51, 52). The analysis indicated USF2 (upstream stimulatory factor 2), a basic helix–loop–helix–leucine–zip transcription factor, as the top predicted upstream transcription factor with 10 of the 18 refractory signature genes enriched in this pathway. USF2 was also the only predicted upstream transcription factor with an adjusted P value < 0.05 (Fig. 5B and SI Appendix, Table S5). We conducted qPCR analysis on USF2 in these RNA samples because it was not included in the CodeSet HuTH17. Short-term (4 h) PMA/ionomycin stimulation dramatically reduced the mRNA levels of USF2 (Fig. 5C). Ionomycin is a calcium ionophore and induces calcium influx in treated cells (53). PMA activates Ca2+-dependent protein kinase C (54). Thus, PMA and ionomycin synergize to activate protein kinase C. Repressed expression of USF2 in PMA/ionomycin-treated T cells indicates that USF2 may be negatively regulated by protein kinase C. We then compared the expression levels of USF2 between responding and nonresponding patients. qPCR analysis showed that the average mRNA level of USF2 was higher in nonresponding patients but not in responding patients compared to healthy controls in the unstimulated cells, while no significant difference was observed in stimulated cells (Fig. 5D). These results suggest an association between enhanced expression of USF2 and increased pathogenic Th17 signaling in nonresponders to anti-TNF therapy in RA, as well as that a strong cell stimulation signal such as PMA/ionomycin stimulation could rapidly shut off or override the function of USF2.
Fig. 5.
The expression of the predicted upstream regulator USF2 is up-regulated in nonresponders. (A) Heat map of the leading-edge genes defined by the GSEA comparisons in Fig. 4D. The enriched genes were categorized into two groups: DE genes unique to NRes (Upper); DE genes shared by Non-Res and Res (Lower). (B) DE genes unique to NRes (n = 18) were subjected to the Enrichr ENCODE TF CHIP-SEq 2015 analysis, and the top five predicted upstream transcription factors ranked by combined score were shown. (C and D) qPCR analysis on USF2 expression in CCR6+ T cells (HC, n = 6; NRes, n = 7; Res, n = 6; unpaired two-tailed Student’s t test, *P < 0.05, mean ± SD). Data are on all samples categorized by with or without stimulation (C) and on stimulated or unstimulated cells categorized by disease diagnosis (D).
USF2-Dependent Proinflammatory Cytokine Expression in Pathogenic Th17 Cells.
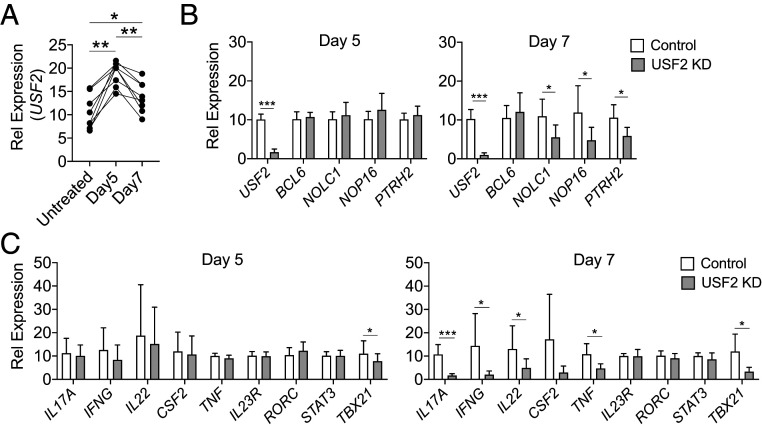
We then tested how USF2 regulates the differentiation of pathogenic Th17 cells induced from naive CD4+ T cells with IL-1β, IL-6, IL-23, and TGF-β cytokine mixture treatment in the presence of anti-CD3/28 stimulation. The levels of USF2 transcripts were comparable among ex vivo isolated peripheral blood mononuclear cells (PBMC), total CD4+ T cells, and naive CD4+ T cells (SI Appendix, Fig. S5A). We observed increased expression of USF2 while subjecting naive and memory CD4+ T cells to the pathogenic Th17 differentiation condition (SI Appendix, Fig. S5B). To investigate whether USF2 regulates pathogenic Th17 development, we used a lentiviral vector to introduce a short hairpin RNA (shRNA) targeting USF2 or a scramble control shRNA into CD4+ T cells during differentiation of pathogenic Th17 cells. In this way, we generated the USF2-knockdown pathogenic Th17 cell lines and their corresponding mock controls. In the control cell lines, we observed an elevated expression of USF2 on days 5 and 7 after subjecting naive CD4 T cells to pathogenic Th17 polarization condition (Fig. 6A). BCL6, NOLC1, NOP16, and PTRH2 are downstream targets of USF2 in human liver cancer cell line HepG2 cells (Fig. 5B and SI Appendix, Table S5), and the expression levels of these genes in nonresponders were elevated compared to healthy controls (Fig. 5A) as well as compared to responders (Fig. 3D). We found, in USF2-knockdown Th17 cells, the expression of NOLC1, NOP16, and PTRH2 but not BCL6 was repressed on day 7 but not on day 5 after subjecting CD4+ T cells to pathogenic Th17 polarization conditions (Fig. 6B), which suggests that 1) although the expression of USF2 was induced at an early stage during pathogenic Th17 differentiation, its regulatory function manifested at a later stage; and 2) the respective USF2-signaling pathways in HepG2 and Th17 cells were likely largely but not completely overlapping. We then looked into Th17-associated cytokines, transcription factors, etc., in USF2-knockdown cells. On day 5, we observed a minor but significant reduction of TBX21 expression (Fig. 6 C, Left). However, after an additional 2 d, not only the reduced expression of TBX21 (P = 0.007) was sustained, the expression of IL17A (P = 0.0004), IFNG (P = 0.029), IL22 (P = 0.024), TNF (P = 0.011), and CSF2 (P = 0.056) was also effectively inhibited without reducing the expression of IL23R, RORC, and STAT3 (Fig. 6 C, Right). These results suggest that USF2 affects T-bet–mediated proinflammatory program and works downstream or independently of ROR-γt, Stat3, and IL-23 receptor during Th17 development. USF2 is critical to sustaining proinflammatory cytokine expression at late stage of Th17 development.
Fig. 6.
Inhibition of USF2 leads to reduced expression of proinflammatory cytokines in Th17 cells. Th17 cells were induced from naive CD4 T cells with anti-CD3/28 treatment in the presence of IL-1β, IL-6, IL-23, and TGF-β1. The expression of USF2 was knocked down in Th17 cells with a lentiviral vector expressing an shRNA targeting USF (USF2 KD) with a lentiviral vector expressing a scramble shRNA as control (Control) in parallel. Gene expression in these cells was assessed by qPCR analysis. (A) Enhanced expression of USF2 in induced Th17 cells. Untreated, untreated nCD4 T cells; Day5 and Day7, 5 and 7 d, respectively, after nCD4 cells were subjected to Th17 polarization condition. HC, n = 8, RM one-way ANOVA, P < 0.0001; multiple comparison test: *P < 0.05; **P < 0.005 (mean ± SD). (B) The sustained expression of NOLC1, NOP16, and PTRH2 but not BCL6 was USF2-dependent. Paired two-tailed Student’s t test, *P < 0.05 and ***P < 0.0005. (C) Inhibition of USF2 reduced the expression of TBX21 and proinflammatory cytokine genes in Th17 cells. HC, n = 8, paired Student’s t test (mean ± SD), *P < 0.05 and ***P < 0.0005. Only comparisons with P < 0.05 were indicated.
Discussion
Through studying the gene expression profiles of CCR6+ T cells from patients responding and not responding to anti-TNF therapy, we found that these cells in nonresponders displayed the gene expression feature of pathogenic Th17 cells, and computational analysis identified transcript factor USF2 as being responsible for this feature. The gene expression of USF2 in CCR6+ T cells from nonresponders was elevated. A previous study has reported USF2-dependent RORC (RORγt in the publication) expression in RORC promoter reporter constructs transfected into HepG2, HeLa, and Jurkat cells and showed that even though small interfering RNA inhibition of USF2 in induced Th17 cells led to reduced expression of RORC, IL17A expression was not reduced (55). However, we found USF2 silencing in induced Th17 cells did not affect RORC expression as well as STAT3 expression, which encodes the key transcription factors RORγt and Stat3 for Th17 differentiation. Instead, USF2 silencing led to reduced gene expression of transcription factor T-bet and proinflammatory cytokines IL-17A, IFN-γ, IL-22, TNF-α, and GM-CSF. Interestingly, the inhibition of TBX21 in USF2-knockdown cultures occurred already on day 5 of the pathogenic Th17 differentiation and preceded the suppression of these effector proinflammatory cytokine genes that only happened at late stage (day 7), which suggests a T-bet–dependent mechanism of prolonged sustainment of proinflammatory cytokines in pathogenic Th17 cells. Taken together, our results suggest that USF2 is dispensable for the initiation of pathogenic Th17 differentiation, but it plays an important role in sustaining inflammatory cytokine expression in pathogenic Th17 cells.
Among the CCR6+ T cells in this study, only about 20% of them secreted IL-17, and high levels of the expression were easily reached upon short-term activation by PMA/ionomycin. CCR6, a CC chemokine receptor protein that belongs to family A of the G protein-coupled receptor superfamily, is not restricted to Th17 cells (26, 56, 57). It is also expressed by a subset of Tregs (58). Thus, these CCR6+ T cells were not only enriched for Th17 cells but likely for CCR6+ Tregs as well. Expression of CCR6 on Tregs facilitates the recruitment of Tregs to inflamed tissue without affecting their suppressive functions (58). Comparing the expression profile of unstimulated CCR6+ T cells of anti-TNF nonresponders to that of responsive patients, we found the elevated expression of LAG3 and CTLA4 (Fig. 3 A and B). LAG-3 and CTLA-4 are expressed by FoxP3+ CD4+ Tregs and mediate Treg suppression. High expression of CTLA-4 positively correlates with suppressive activity of Tregs (40). Hence, the increased expression of LAG3 and CTLA4 in responders suggests higher Treg activity in patients responsive to anti-TNF therapy compared to nonresponders.
Consistent with this observation, the IPA upstream regulator analysis using the differentially expressed genes in CCR6+ T cells between responding vs. nonresponding patients showed that the butyric acid and curcumin pathways were down-regulated in nonresponding patients. Butyric acid is a microbial metabolite found in many food products, especially milk products. Butyrate, the salt format of butyric acid, promotes Treg generation and inhibits Th17 differentiation and is found to ameliorate autoimmune uveitis and colitis in animal models (46, 47, 59). Curcumin is a chemical extracted from Curcuma longa plants used in cooking and cosmetics. It has antioxidant and antiinflammatory properties (49). Curcumin has been shown to inhibit Th17 differentiation both in animal models (60) and in in vitro human studies (61). On the other hand, multiple proinflammatory pathways including the IL-1β, NF-κB, and GM-CSF–signaling pathways were activated in nonresponding patients.
Thus, our results showed inadequate suppression of proinflammatory immune responses by anti-TNF in refractory patients, and the USF2-signaling pathway may be responsible for the resistance to treatment. USF2 is ubiquitously expressed and participates in embryonic development, brain function, metabolism (62), and possibly in hematopoietic stem cell development (63). Several studies suggest that USF2 also contributes to cancer development (64, 65). Yet little is known about the function of USF2 in regulating the immune system, not to mention in the context of pathogenic Th17 regulation. Further study of the role of USF2 in pathogenic Th17 function may shed a light on disease pathogenesis and lead to development of novel therapeutic interventions for refractory RA.
Materials and Methods
Human Subjects.
Blood samples were obtained from 6 healthy donors and 14 patients with established RA. The study was approved by the Regional Ethics Board in Gothenburg, Sweden (Dnr 633-07), and informed written consent was obtained from all participants. The patients fulfilled the American College of Rheumatology 1987 revised criteria for RA (66). One of the 14 patients was excluded from the study due to inadequate in vitro stimulation for gene expression analysis. All of the remaining 13 RA patients were female and obtained infliximab injection intravenously (i.v.) at a dose of 200 mg every 8 wk. Twelve patients were also treated with methotrexate (7.5–25 mg/wk), and the remaining patient was treated with mycophenolate mofetil (2 g/d). Response to treatment was determined according to DAS28 criteria, with DAS28 < 2.8 as responding and ≥ 2.8 as nonresponding at the time of blood sampling. All of the patients were refractory to treatment with conventional disease-modifying drugs, such as methotrexate and combination of methotrexate with hydroxychloroquine. Two of the patients were previously treated with a different anti-TNF drug (etanercept). Six patients responded to treatment including the mycophenolate mofetil-treated one, and the other seven were nonresponders. Four patients of each group were ACPA-positive. Mean total dose of infliximab for the responders and nonresponders were 8,367 and 8,257 mg, respectively. The control group comprised six healthy subjects with no report of any autoimmune disease or the use of any pharmacological drug. Blood samples from healthy donors were also obtained from the Partners Multiple Sclerosis Center at Brigham and Women’s Hospital under Institutional Review Board (IRB) Protocol 1999P010435 for cell surface antigen and intracellular cytokine-staining assays and in vitro Th17 cell differentiation analysis.
Cell Isolation and Stimulation.
PBMC were isolated with Ficoll-Paque PLUS (GE Healthcare) separation. Total CD4+ T cells were isolated from PBMC with the EasySep Human CD4+ T Cell Enrichment Kit (StemCell Technologies, catalog number 19052) following manufacturer’s instruction. Naive CD4+ T cells were isolated from total CD4+ T cells with the EasySep Human Naive CD4+ T Cell Isolation Kit (StemCell Technologies, catalog number 19555) following manufacturer’s instruction. CCR6+ T cells were isolated from PBMC with the EasySep Human Th17 Cell Enrichment Kit II (StemCell Technologies, catalog number 18162) following manufacturer’s instruction as previously described (31). Briefly, PBMC were treated with EasySep Human CD4+CXCR3− T Cell Pre-Enrichment Cocktail, which removes cells expressing CD8, CD14, CD16, CD19, CD20, CD36, CD56, CD66b, CD123, TCRγ/δ, glycophorin A, CD45RA, and CXCR3 by negative selection. Then CCR6+ cells were selected using EasySep Human CCR6 Positive Selection Cocktail. Resulting CD4+CCR6+CXCR3− T cells were resuspended in Roswell Park Memorial Institute (RPMI) 1640 medium supplemented with heat-inactivated 10% fetal calf serum and seeded in a U-bottom 96-well plate at 1–2 × 106 cells/100 μL/well. Equal volume of medium with or without PMA (60 ng/mL) and ionomycin (1 μg/mL) was added to each well. Cells were incubated at 37 °C, 5% CO2 for 4 h. Cells were then pelleted and lysed in 100 μL of Buffer RLT Plus containing 1 μL of β-mercaptoethanol (Qiagen, catalog number 1053393) following manufacturer’s instruction. Cell lysates were stored at −80 °C for later gene expression analysis.
Cell Surface Antigen and Intracellular Cytokine-Staining Assays.
Total memory CD4+ T cells were isolated from PBMC by negative selection using the EasySep Human memory CD4+ T Cell Enrichment Kit (StemCell Technologies, catalog number 19157) following manufacturer’s instruction, and CCR6+ T cells were isolated as described in Cell Isolation and Stimulation. For cell surface antigen staining, isolated cells were seeded into a U-bottom 96-well plate (up to 1 × 105 cells per well) and stained with anti–CD3-Brilliant Violet 605 (clone: OKT3, Biolegend), anti–CD4-pacific blue (clone: RPA-T4, BD Biosciences), anti–CD8-Alexa Fluor 700 (clone: HIT8a, BioLegend), anti–CD19-FITC (clone: HIB19, BioLegend), anti–CCR4-PE (clone: 1G1, BD Biosciences), anti–CCR6-Alexa Fluor 647 (clone: G034E3, BioLegend), and anti–CXCR3-PerCP/Cy5.5 (clone: G025H7, BioLegend). Cell surface antigen staining on CD4+ lymphocytes was accessed with FlowJo. For intracellular cytokine staining, assays were carried out with staining buffers and antibodies from BD Biosciences as described before (19). Briefly, isolated cells were seeded into a U-bottom 96-well plate (up to 1 × 105 cells per well) and stimulated with PMA (100 ng/mL) and ionomycin (1 μg/mL) in the presence of GolgiStop (catalog number 554724) for 4 h. Cells were then fixed with BD Cytofix fixation buffer (catalog number 554655) and washed with BD Perm/Wash buffer (catalog number 554723) following manufacturer’s instruction. Cells in each well were equally divided and seeded into two new wells, with one for intracellular cytokine staining and the other for isotype control staining. The following fluorophore-conjugated antibodies from BD Biosciences were used for staining analysis or as isotype controls: anti–CD4-pacific blue (clone: RPA-T4), anti–IL-17A-Alexa Fluor 647 (clone: N49-653), anti–IFN-γ–FITC (clone: B27), anti–IL-10-PE (clone: JES3-19F), mouse IgG1-Alexa Fluor 647 (clone: MOPC-21), mouse IgG1-FITC (clone: MOPC-21), and rat IgG2a-PE (clone: R35-95). Stained cells were analyzed with a BD LSR II cytometer. Cytokine secretion in CD4+ lymphocytes was accessed with FlowJo.
In Vitro Knockdown with shRNA Lentivirus in Th17 Cells.
On day 0, naive CD4+ T cells (30,000 cells) were cultured in a 96-well plate in X-VIVO 15 Serum-free Hematopoietic Cell Medium (Lonza Pharma & Biotech, catalog number 04-418Q) in the presence of STEMCELL ImmunoCult Human CD3/CD28 (StemCell Technologies, catalog number 10971). At 24 h, a lentiviral vector carrying a USF2-targeting shRNA (GeneCopoeia, LPP-CS-HSH111132-shM03-01-100) was added to the culture at multiplicity of infection of 5 to knock down the expression of USF2. A lentiviral vector carrying a scramble shRNA (GeneCopoeia, LPP-CSHCTR001-shM03-300) was used as a control. The lentiviral vectors also expressed the enhanced green fluorescent protein (eGFP) reporter gene and puromycin resistant gene. At 48 h, cells were subjected to Th17 polarization condition: IL-1β (R&D, #201-LB-005) at 12.5 ng/mL, IL-6 (R&D, #206-IL-010) at 25 ng/mL, IL-23 (R&D, #1290-IL-010) at 25 ng/mL, TGF-β (R&D, #240-B-002) at 5 ng/mL, anti–IFN-γ (Clone B27) (BD Biosciences, #554698) at 1 µg/mL, and anti–IL-4 (Clone 34019) (R&D, #MAB204-100) at 1 µg/mL. On day 4, cells were treated with puromycin (Fisher Scientific, catalog number A1113803) at 1 µg/mL On days 7 and 9 (Th17 polarization days 5 and 7), cells were stained with propidium iodide (BioLegend, 421301) at 10 µg/mL, and GFP+ live cells were sorted by a BD FACSAria sorter for RNA isolation.
Low-Input Quantitative Real-Time PCR.
Total RNA in cells (PBMC, CD4+ T cells, Th17 cells, and CCR6+ T cells) was isolated with the Qiagen RNeasy Plus Mini Kit (catalog number 74234). Complementary DNA (cDNA) was synthesized with the SuperScript VILO master mix (catalog number 11755050) and preamplified for 14 cycles with the TaqMan preAmp master mix (catalog number 4391128) following the manufacturer’s instruction. qPCR analysis was run and analyzed with the ViiA 7 Real-Time PCR System (Life Technologies) using the TaqMan fast universal PCR master mix (2x) (catalog number 4352042) and qPCR primers (SI Appendix, Table S6) purchased from ThermoFisher Scientific. The comparative threshold cycle method and an internal control (β2m) were used for normalization of the target genes. Relative expression was calculated as: ΔCT = CTgene of interest − CTβ2m; ΔΔCT = ΔCT sample of interest − mean of ΔCT healthy control; the relative change of gene expression between the expression level of sample of interest and the mean expression level of healthy controls was given by this formula: (2−ΔΔCT) × 10. All qPCR reactions were performed in duplicate.
nCounter Analysis of Gene Expression.
As previously described, we designed a nanoString CodeSet HuTH17 that constitutes a 418-gene expression detection panel specific for human T cell activation and differentiation (19). Cell lysates prepared as described above from stimulated and unstimulated isolated CCR6+CD4+ cells were subjected to the nCounter Gene Expression Analysis using the CodeSet HuTH17 according to the protocol provided by the manufacturer (NanoString Technologies).
Data Analysis.
Similar to the approaches we described previously (19), nCounter gene expression data were normalized for code count using the geometric mean, for background using the mean, and for sample content using the geometric mean of housekeeping genes (B2M, RPL3, and beta actin) with R (version 3.2.0) and NanoStringNorm (version 1.2.1). Mean plus 2 SDs of detected expression values of negative controls of the CodeSet HuTH17 was used as the cutoff to select for expressed genes, which have the minimum expression value across all samples in an unbiased manner above the cutoff value. Using this criterion, 397 of the 415 genes (excluding the three housekeeping genes) were selected as expressed genes and subjected to further analyses. Gene expression heat maps were generated with GENE-E (https://software.broadinstitute.org/GENE-E/) using color-value as the z-scores of a data point. PCA plots were generated using R, factoextra (v1.0.5), and ggplot (v2.2.1). As described previously (19), mouse gene expression data were downloaded from the Gene Expression Omnibus (GEO) (GSE39820) (18) and normalized using Robust Multi-array Average (RMA) (67) and ComBat (68) in GenePattern (https://www.genepattern.org/), and genes with multiple probes were collapsed to unique genes by selecting the probe with the highest average expression across all samples. GSEA was done in GenePattern using default settings (weighted scoring scheme, Signal2Noise metric, 1,000 permutations) (69–71) to test the enrichment of human signatures in the mouse expression profiles. Storey’s q-value is used to control the false discovery rate. The IPA canonical pathway analysis and upstream regulator analysis were performed for the differentially expressed genes (using corresponding fold changes and P values) to identify key upstream regulatory molecules. Canonical pathways and upstream regulators with z-scores ≥ 2 and z-scores ≤ −2 were defined as activator and inhibitor mechanisms, respectively. IPA was also used to generate the network diagram. In addition, Enrichr (https://maayanlab.cloud/Enrichr/) was also utilized to conduct transcriptional and pathway analysis, in which the P value is computed from the Fisher exact test; the adjusted P value is a rank-based ranking derived from running the Fisher exact test for many random gene sets to compute a mean rank and SD from the expected rank for each term in the gene set library. The z-score is defined as the deviation from the expected rank, and the combined score is calculated by log of the P value multiplied by the z-score.
Statistical Analysis.
Paired Student’s t test was performed to compare the T cell subset frequencies. Unpaired Student’s t test was performed to compare differential gene expression between responders vs. nonresponders, responders vs. HC, and nonresponders vs. HC, respectively, with Excel. Two-tailed P values < 0.05 were considered statistically significant. Statistical analysis was performed with Prism 7 and 8 (GraphPad Software), and one-way ANOVA was performed with R statistical software (version 3.2.0) to compare the differential gene expression among healthy controls and responding and nonresponding patients with RA. Genes with P value < 0.05 were selected for PCA (Fig. 2 C and D).
Supplementary Material
Acknowledgments
We wish to thank Ioannis Vlachos for scientific discussion. This work was supported in part by NIH P01 Grant NS076410 (to H.L.W.), National Multiple Sclerosis Society Research Grant RG-1507-05029 (to H.L.W.), National Multiple Sclerosis Society Research Grant RG-1907-34563 (to D.H.), Swedish Research Council Grant 521-2014-2637 (to M.I.B.), and the regional agreement on medical training and clinical research between the Western Götaland county council and the University of Göteborg Grant ALFGBG-671631 (to M.I.B.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2007935117/-/DCSupplemental.
Data Availability.
The main data supporting the findings of this study are available within the article and SI Appendix.
References
- 1.Cope A. P., Schulze-Koops H., Aringer M., The central role of T cells in rheumatoid arthritis. Clin. Exp. Rheumatol. 25, S4–S11 (2007). [PubMed] [Google Scholar]
- 2.Miossec P., Interleukin-17 in rheumatoid arthritis: If T cells were to contribute to inflammation and destruction through synergy. Arthritis Rheum. 48, 594–601 (2003). [DOI] [PubMed] [Google Scholar]
- 3.Noack M., Miossec P., Selected cytokine pathways in rheumatoid arthritis. Semin. Immunopathol. 39, 365–383 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Yago T., et al. , IL-23 and Th17 disease in inflammatory arthritis. J. Clin. Med. 6, 81 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kalliolias G. D., Ivashkiv L. B., TNF biology, pathogenic mechanisms and emerging therapeutic strategies. Nat. Rev. Rheumatol. 12, 49–62 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Evans H. G., et al. , In vivo activated monocytes from the site of inflammation in humans specifically promote Th17 responses. Proc. Natl. Acad. Sci. U.S.A. 106, 6232–6237 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Perdomo-Celis F., Salgado D. M., Narváez C. F., Selective dysfunction of subsets of peripheral blood mononuclear cells during pediatric dengue and its relationship with clinical outcome. Virology 507, 11–19 (2017). [DOI] [PubMed] [Google Scholar]
- 8.Ma X., Xu S., TNF inhibitor therapy for rheumatoid arthritis. Biomed. Rep. 1, 177–184 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Langrish C. L., et al. , IL-23 drives a pathogenic T cell population that induces autoimmune inflammation. J. Exp. Med. 201, 233–240 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Park H., et al. , A distinct lineage of CD4 T cells regulates tissue inflammation by producing interleukin 17. Nat. Immunol. 6, 1133–1141 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Waite J. C., Skokos D., Th17 response and inflammatory autoimmune diseases. Int. J. Inflamm. 2012, 819467 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Patel D. D., Kuchroo V. K., Th17 cell pathway in human immunity: Lessons from genetics and therapeutic interventions. Immunity 43, 1040–1051 (2015). [DOI] [PubMed] [Google Scholar]
- 13.Shen H., Goodall J. C., Hill Gaston J. S., Frequency and phenotype of peripheral blood Th17 cells in ankylosing spondylitis and rheumatoid arthritis. Arthritis Rheum. 60, 1647–1656 (2009). [DOI] [PubMed] [Google Scholar]
- 14.Alunno A., et al. , Altered immunoregulation in rheumatoid arthritis: The role of regulatory T cells and proinflammatory Th17 cells and therapeutic implications. Mediators Inflamm. 2015, 751793 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shen H., Xia L., Lu J., Xiao W., Infliximab reduces the frequency of interleukin 17-producing cells and the amounts of interleukin 17 in patients with rheumatoid arthritis. J. Investig. Med. 58, 905–908 (2010). [DOI] [PubMed] [Google Scholar]
- 16.Hirota K., et al. , Autoimmune Th17 cells induced synovial stromal and innate lymphoid cell secretion of the cytokine GM-CSF to initiate and augment autoimmune arthritis. Immunity 48, 1220–1232.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McGeachy M. J., et al. , TGF-beta and IL-6 drive the production of IL-17 and IL-10 by T cells and restrain T(H)-17 cell-mediated pathology. Nat. Immunol. 8, 1390–1397 (2007). [DOI] [PubMed] [Google Scholar]
- 18.Lee Y., et al. , Induction and molecular signature of pathogenic TH17 cells. Nat. Immunol. 13, 991–999 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu D., et al. , Transcriptional signature of human pro-inflammatory TH17 cells identifies reduced IL10 gene expression in multiple sclerosis. Nat. Commun. 8, 1600 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Colombel J. F., Sendid B., Jouault T., Poulain D., Secukinumab failure in Crohn’s disease: The yeast connection? Gut 62, 800–801 (2013). [DOI] [PubMed] [Google Scholar]
- 21.Hueber W. et al.; Secukinumab in Crohn’s Disease Study Group , Secukinumab, a human anti-IL-17a monoclonal antibody, for moderate to severe Crohn’s disease: Unexpected results of a randomised, double-blind placebo-controlled trial. Gut 61, 1693–1700 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fragoulis G. E., Siebert S., McInnes I. B., Therapeutic targeting of IL-17 and IL-23 cytokines in immune-mediated diseases. Annu. Rev. Med. 67, 337–353 (2016). [DOI] [PubMed] [Google Scholar]
- 23.Cosmi L., et al. , Evidence of the transient nature of the Th17 phenotype of CD4+CD161+ T cells in the synovial fluid of patients with juvenile idiopathic arthritis. Arthritis Rheum. 63, 2504–2515 (2011). [DOI] [PubMed] [Google Scholar]
- 24.Nistala K., et al. , Th17 plasticity in human autoimmune arthritis is driven by the inflammatory environment. Proc. Natl. Acad. Sci. U.S.A. 107, 14751–14756 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kotake S., Yago T., Kobashigawa T., Nanke Y., The plasticity of Th17 cells in the pathogenesis of rheumatoid arthritis. J. Clin. Med. 6, 67 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sallusto F., Zielinski C. E., Lanzavecchia A., Human Th17 subsets. Eur. J. Immunol. 42, 2215–2220 (2012). [DOI] [PubMed] [Google Scholar]
- 27.Paulissen S. M., van Hamburg J. P., Dankers W., Lubberts E., The role and modulation of CCR6+ Th17 cell populations in rheumatoid arthritis. Cytokine 74, 43–53 (2015). [DOI] [PubMed] [Google Scholar]
- 28.Zheng Y., et al. , TNFα promotes Th17 cell differentiation through IL-6 and IL-1β produced by monocytes in rheumatoid arthritis. J. Immunol. Res. 2014, 385352 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang C. Q. F., et al. , IL-17 and TNF synergistically modulate cytokine expression while suppressing melanogenesis: Potential relevance to psoriasis. J. Invest. Dermatol. 133, 2741–2752 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gabr M. A., et al. , Interleukin-17 synergizes with IFNγ or TNFα to promote inflammatory mediator release and intercellular adhesion molecule-1 (ICAM-1) expression in human intervertebral disc cells. J. Orthop. Res. 29, 1–7 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Andersson K. M., et al. , Pathogenic transdifferentiation of Th17 cells contribute to perpetuation of rheumatoid arthritis during anti-TNF treatment. Mol. Med. 21, 536–543 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Glatt S., et al. , Efficacy and safety of bimekizumab as add-on therapy for rheumatoid arthritis in patients with inadequate response to certolizumab pegol: A proof-of-concept study. Ann. Rheum. Dis. 78, 1033–1040 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Buch M. H., Bingham S. J., Bryer D., Emery P., Long-term infliximab treatment in rheumatoid arthritis: Subsequent outcome of initial responders. Rheumatology (Oxford) 46, 1153–1156 (2007). [DOI] [PubMed] [Google Scholar]
- 34.Rubbert-Roth A., Finckh A., Treatment options in patients with rheumatoid arthritis failing initial TNF inhibitor therapy: A critical review. Arthritis Res. Ther. 11 (suppl. 1), S1 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hirota K., et al. , Preferential recruitment of CCR6-expressing Th17 cells to inflamed joints via CCL20 in rheumatoid arthritis and its animal model. J. Exp. Med. 204, 2803–2812 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Singh S. P., Zhang H. H., Foley J. F., Hedrick M. N., Farber J. M., Human T cells that are able to produce IL-17 express the chemokine receptor CCR6. J. Immunol. 180, 214–221 (2008). [DOI] [PubMed] [Google Scholar]
- 37.Groom J. R., Luster A. D., CXCR3 in T cell function. Exp. Cell Res. 317, 620–631 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wherry E. J., et al. , Molecular signature of CD8+ T cell exhaustion during chronic viral infection. Immunity 27, 670–684 (2007). [DOI] [PubMed] [Google Scholar]
- 39.McKinney E. F., Lee J. C., Jayne D. R., Lyons P. A., Smith K. G., T-cell exhaustion, co-stimulation and clinical outcome in autoimmunity and infection. Nature 523, 612–616 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen X., Oppenheim J. J., Resolving the identity myth: Key markers of functional CD4+FoxP3+ regulatory T cells. Int. Immunopharmacol. 11, 1489–1496 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Macian F., NFAT proteins: Key regulators of T-cell development and function. Nat. Rev. Immunol. 5, 472–484 (2005). [DOI] [PubMed] [Google Scholar]
- 42.Serfling E., et al. , NFAT transcription factors in control of peripheral T cell tolerance. Eur. J. Immunol. 36, 2837–2843 (2006). [DOI] [PubMed] [Google Scholar]
- 43.Di Salvo E., Ventura-Spagnolo E., Casciaro M., Navarra M., Gangemi S., IL-33/IL-31 axis: A potential inflammatory pathway. Mediators Inflamm. 2018, 3858032 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Eidenschenk C., Rutz S., Liesenfeld O., Ouyang W., Role of IL-22 in microbial host defense. Curr. Top. Microbiol. Immunol. 380, 213–236 (2014). [DOI] [PubMed] [Google Scholar]
- 45.Menezes M. E., et al. , Role of MDA-7/IL-24 a multifunction protein in human diseases. Adv. Cancer Res. 138, 143–182 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Arpaia N., et al. , Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature 504, 451–455 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen X., et al. , Sodium butyrate regulates Th17/Treg cell balance to ameliorate uveitis via the Nrf2/HO-1 pathway. Biochem. Pharmacol. 142, 111–119 (2017). [DOI] [PubMed] [Google Scholar]
- 48.Griffiths K., et al. , Food antioxidants and their anti-inflammatory properties: A potential role in cardiovascular diseases and cancer prevention. Diseases 4, 28 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hewlings S. J., Kalman D. S., Curcumin: A review of its effects on human health. Foods 6, 92 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brück J., et al. , Nutritional control of IL-23/Th17-mediated autoimmune disease through HO-1/STAT3 activation. Sci. Rep. 7, 44482 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen E. Y., et al. , Enrichr: Interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 14, 128 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kuleshov M. V., et al. , Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 44, W90–W97 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chatila T., Silverman L., Miller R., Geha R., Mechanisms of T cell activation by the calcium ionophore ionomycin. J. Immunol. 143, 1283–1289 (1989). [PubMed] [Google Scholar]
- 54.Castagna M., et al. , Direct activation of calcium-activated, phospholipid-dependent protein kinase by tumor-promoting phorbol esters. J. Biol. Chem. 257, 7847–7851 (1982). [PubMed] [Google Scholar]
- 55.Ratajewski M., Walczak-Drzewiecka A., Salkowska A., Dastych J., Upstream stimulating factors regulate the expression of RORγT in human lymphocytes. J. Immunol. 189, 3034–3042 (2012). [DOI] [PubMed] [Google Scholar]
- 56.Rot A., von Andrian U. H., Chemokines in innate and adaptive host defense: Basic chemokinese grammar for immune cells. Annu. Rev. Immunol. 22, 891–928 (2004). [DOI] [PubMed] [Google Scholar]
- 57.Ito T., Carson W. F. 4th, Cavassani K. A., Connett J. M., Kunkel S. L., CCR6 as a mediator of immunity in the lung and gut. Exp. Cell Res. 317, 613–619 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kleinewietfeld M., et al. , CCR6 expression defines regulatory effector/memory-like cells within the CD25(+)CD4+ T-cell subset. Blood 105, 2877–2886 (2005). [DOI] [PubMed] [Google Scholar]
- 59.Zhang M., et al. , Butyrate inhibits interleukin-17 and generates Tregs to ameliorate colorectal colitis in rats. BMC Gastroenterol. 16, 84 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhao G., et al. , Curcumin inhibiting Th17 cell differentiation by regulating the metabotropic glutamate receptor-4 expression on dendritic cells. Int. Immunopharmacol. 46, 80–86 (2017). [DOI] [PubMed] [Google Scholar]
- 61.Handono K., Pratama M. Z., Endharti A. T., Kalim H., Treatment of low doses curcumin could modulate Th17/Treg balance specifically on CD4+ T cell cultures of systemic lupus erythematosus patients. Cent. Eur. J. Immunol. 40, 461–469 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Horbach T., Götz C., Kietzmann T., Dimova E. Y., Protein kinases as switches for the function of upstream stimulatory factors: Implications for tissue injury and cancer. Front. Pharmacol. 6, 3 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Belew M. S., et al. , PLAG1 and USF2 Co-regulate expression of musashi-2 in human hematopoietic stem and progenitor cells. Stem Cell Rep. 10, 1384–1397 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chi T. F., Horbach T., Götz C., Kietzmann T., Dimova E. Y., Cyclin-dependent kinase 5 (CDK5)-mediated phosphorylation of upstream stimulatory factor 2 (USF2) contributes to carcinogenesis. Cancers (Basel) 11, 523 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tan Y., Chen Y., Du M., Peng Z., Xie P., USF2 inhibits the transcriptional activity of Smurf1 and Smurf2 to promote breast cancer tumorigenesis. Cell. Signal. 53, 49–58 (2019). [DOI] [PubMed] [Google Scholar]
- 66.Arnett F. C., et al. , The American Rheumatism Association 1987 revised criteria for the classification of rheumatoid arthritis. Arthritis Rheum. 31, 315–324 (1988). [DOI] [PubMed] [Google Scholar]
- 67.Irizarry R. A., et al. , Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4, 249–264 (2003). [DOI] [PubMed] [Google Scholar]
- 68.Johnson W. E., Li C., Rabinovic A., Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 8, 118–127 (2007). [DOI] [PubMed] [Google Scholar]
- 69.Reich M., et al. , GenePattern 2.0. Nat. Genet. 38, 500–501 (2006). [DOI] [PubMed] [Google Scholar]
- 70.Subramanian A., et al. , Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mootha V. K., et al. , PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 34, 267–273 (2003). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The main data supporting the findings of this study are available within the article and SI Appendix.