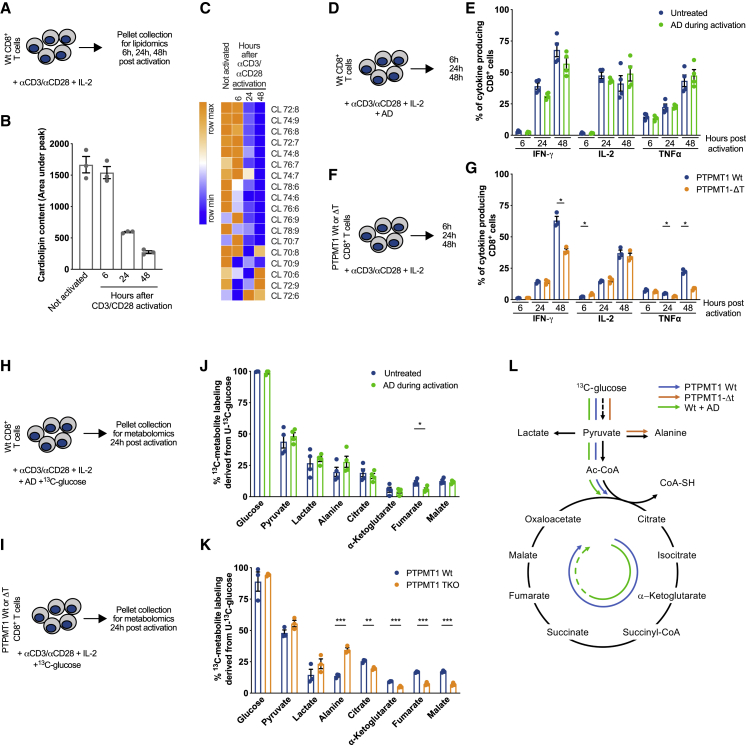
Figure 3.
Metabolic Reprogramming upon CD8+ T Cell Activation Requires Cardiolipin, but Not Its De Novo Synthesis
(A) Schematic of CD8+ T cells activated with αCD3/αCD28 +IL-2. At indicated time points cell pellets were collected for lipidomics analysis.
(B) Total CL amount in CD8+ T cells treated as in (A)
(C) Cardiolipin species in CD8+ T cells treated as in (A).
(D) Schematic of WT CD8+ T cells activated with αCD3/αCD28 +IL-2 with ± AD.
(E) Cytokine production of WT CD8+ T cells activated as in (D) analyzed at indicated time points.
(F) Schematic of WT and Ptpmt1−/− CD8+ T cells activated with αCD3/αCD28 + IL-2.
(G) Cytokine production of WT and Ptpmt1−/− CD8+ T cells activated as in (F) analyzed at indicated time points.
(H) WT CD8+ T cells activated with αCD3/αCD28 + IL-2 in a medium containing 10mM 13C-glucose ± AD. Twenty-four h after activation cell pellets were collected for metabolomics analysis.
(I) WT and Ptpmt1−/− CD8+ T cells activated with αCD3/αCD28 + IL-2 in a medium containing 10mM 13C-glucose. Twenty-four h after activation cell pellets were collected for metabolomics analysis.
(J) 13C-glucose derived carbons incorporated into indicated metabolites in WT CD8+ T cells treated as in (H). Data represent mean ± SEM of 3 samples per genotype.
(K) 13C-glucose derived carbons incorporated into indicated metabolites in Ptpmt1 WT and Ptpmt1−/− CD8+ T cells treated as in (I). Data represent mean ± SEM of 3 samples per genotype.
(L) Schematic of 13C-glucose tracing experiment results in WT CD8+ T cells ± AD, Ptpmt1 WT, and Ptpmt1−/− CD8+ T cells.
Data shown as mean ± SEM of ≥3 independent experiments unless indicated. Statistical comparisons for 2 groups calculated by unpaired two-tailed Student’s t test, ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001.
See also Figure S3.