Fig. 1 |. Schematics of the GRID-seq experiment.
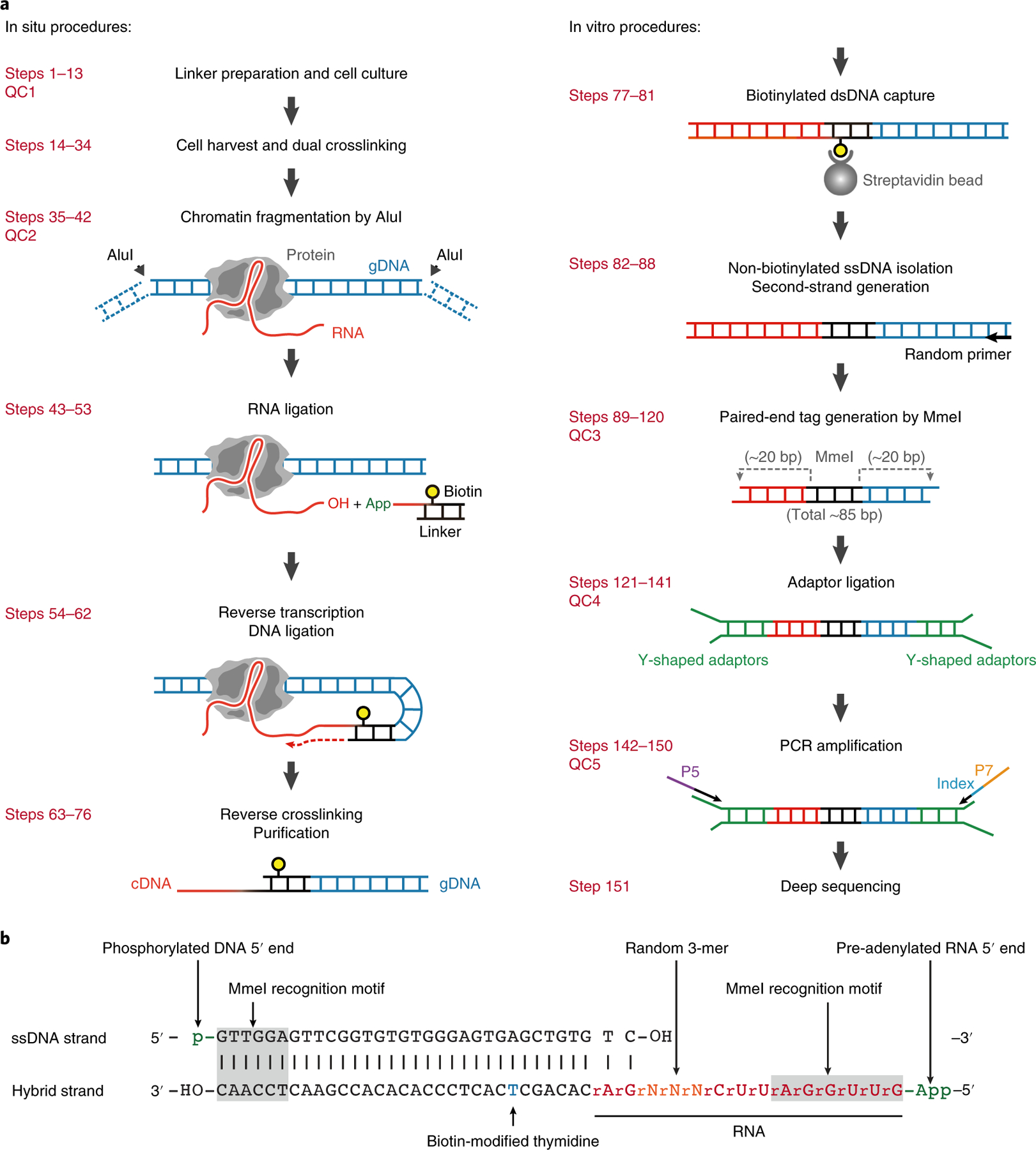
a, Left, GRID-seq experimental steps performed in situ on fixed nuclei. A bivalent linker is used to ligate chromatin-associated RNA (with the ssRNA stretch on the linker) and fragmented genomic DNA crosslinked in proximity. Right, steps performed in solution. b, The structure of the bivalent GRID-seq Linker. The top strand is a 5′-phosphorylated DNA sequence (black) and the bottom strand is a hybrid consisting of both DNA and RNA bases (red) with a biotin-modified thymidine (blue). Both ends of the linker carry an MmeI restriction site (gray-shaded). App, pre-adenylation.
