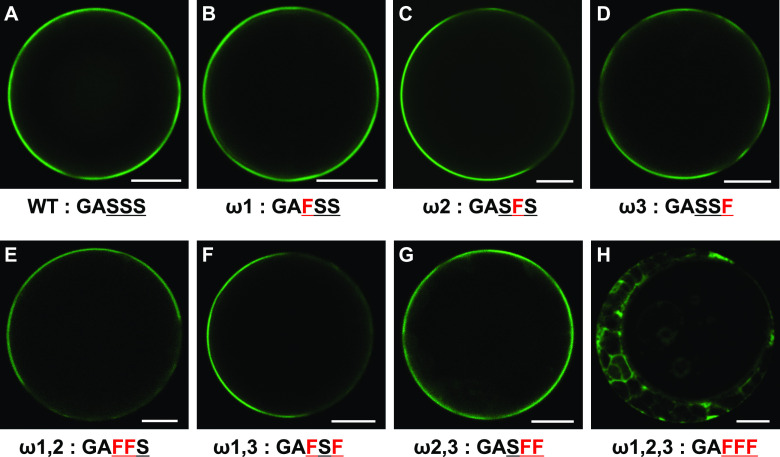
Figure 7.
Analysis of the EPAD1 GPI-Anchor Sites.
Representative GFP fluorescence images of Arabidopsis protoplast cells expressing 35Spro:SP-GFP-cEPAD1 wild type (WT; A), 35Spro:SP-GFP-cEPAD1 ω1 (B), 35Spro:SP-GFP-cEPAD1 ω2 (C), 35Spro:SP-GFP-cEPAD1 ω3 (D), 35Spro:SP-GFP-cEPAD1 ω1,2 (E), 35Spro:SP-GFP-cEPAD1 ω1,3 (F), 35Spro:SP-GFP-cEPAD1 ω2,3 (G), 35Spro:SP-GFP-cEPAD1 ω1,2,3 (H). Amino acid sequence under each panel indicates the position of the mutation in the GPI-anchor attachment sequence in red. Underlined residues are predicted GPI-anchor attachment sites. At least ten randomly chosen protoplasts were used for GFP signal analysis. Representative images are shown. Bars = 10 µm.