Figure 1.
Identification of the Dimerization Domain of DDX21
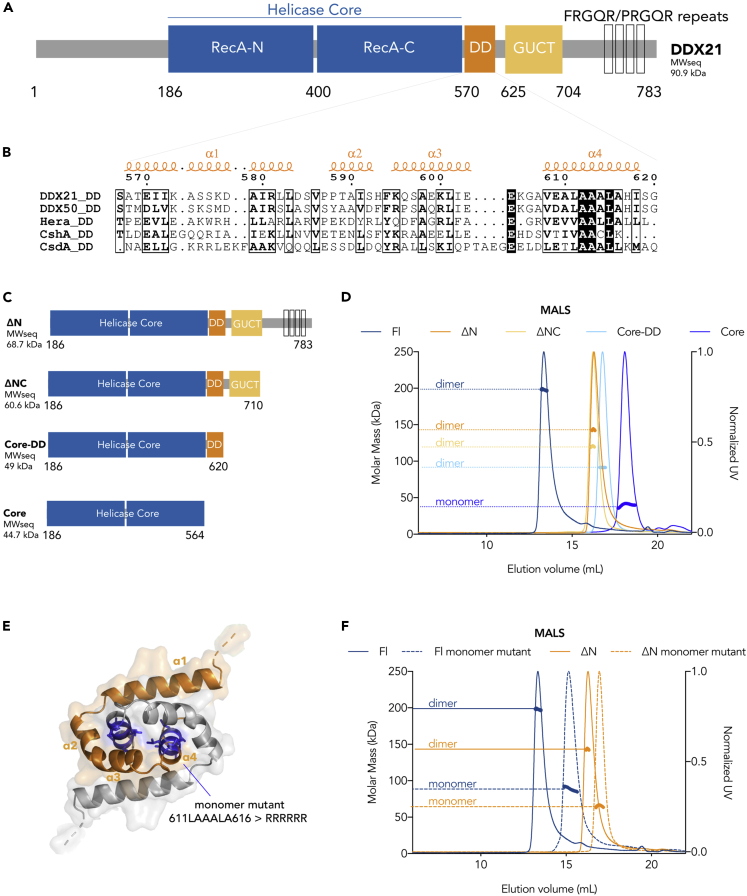
(A) Numbered linear sequence with the different domains colored in blue (HC), orange (DD), and yellow (GUCT) and the C-terminal tail with the FRGQR/PRGQR repeats in black boxes. The molecular weight of this sequence is MWseq = 90.9 kDa (cleaved affinity tag) (see also Figures S1, Figure 1, Figure 2, Figure 3, Figure 4, Figure 5and S7).
(B) Sequence alignment of the DDs from the two human paralogs DDX21 and DDX50, as well as the bacterial dimeric DEAD-box helicases Hera, CshA, and CsdA (see also Figure S3).
(C) Schematic representation of the truncation constructs used in this study with their corresponding monomeric MWseq calculated from their sequences.
(D) SEC-MALS data show that all the constructs except DDX21Core are dimers (theoretical molar masses [MWseq] for Fl, ΔN, ΔNC, Core-DD, and Core are 90.9, 68.7, 60.6, 49, and 44.7 kDa, respectively). Experimental SEC-MALS molar masses are Fl, 198 kDa; ΔN, 143 kDa; ΔNC, 120 kDa; Core-DD, 91 kDa; and Core, 41 kDa) (see also Figures S4 and S5).
(E) Homology model of the DD of DDX21 in cartoon representation, with helices α1 to α4 of each protomer colored in orange and gray, respectively. The residues in α4 mutated to create the monomeric mutants are shown in blue stick representation.
(F) SEC-MALS of the Fl and ΔN monomeric mutants (in dashed lines) with molar masses of 88 and 66 kDa, compared with intact DDX21Fl (blue) and DDX21ΔN (orange) with molar masses of 198 and 143 kDa.