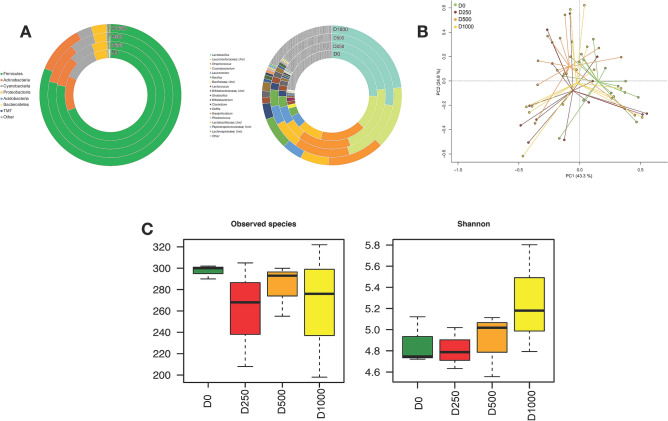
Figure 3.
Autochthonous intestinal microbiota from sea bass Dicentrarchus labrax fed with control diet (D0) and different doses of microencapsulated OA and NIC (D250, D500 and D1000). (A) Average phylogenetic profiles of the gut microbiome of sea bass are provided at phylum (left) and genus (right) level; Phyla and genus which represent < 1.5% of the autochthonous community were included in ‘Others’. (B) Principal Component Analysis (PCA) of the Euclidean distance of sea bass gut microbial ecosystem after the assumption of different diets (D0, D250, D500 and D1000). A significant separation among groups was observed [P value = 0.03; Permutation test with pseudo-F ratios (Adonis)]. (C) Alpha diversity of the sea bass GM for all diet assumption was estimated with observed species and Shannon metrics.