Fig. 8. Bypass mutants lacking channel proteins differ in cleavage of Pro-σK and SpoIVFB-GFP localization.
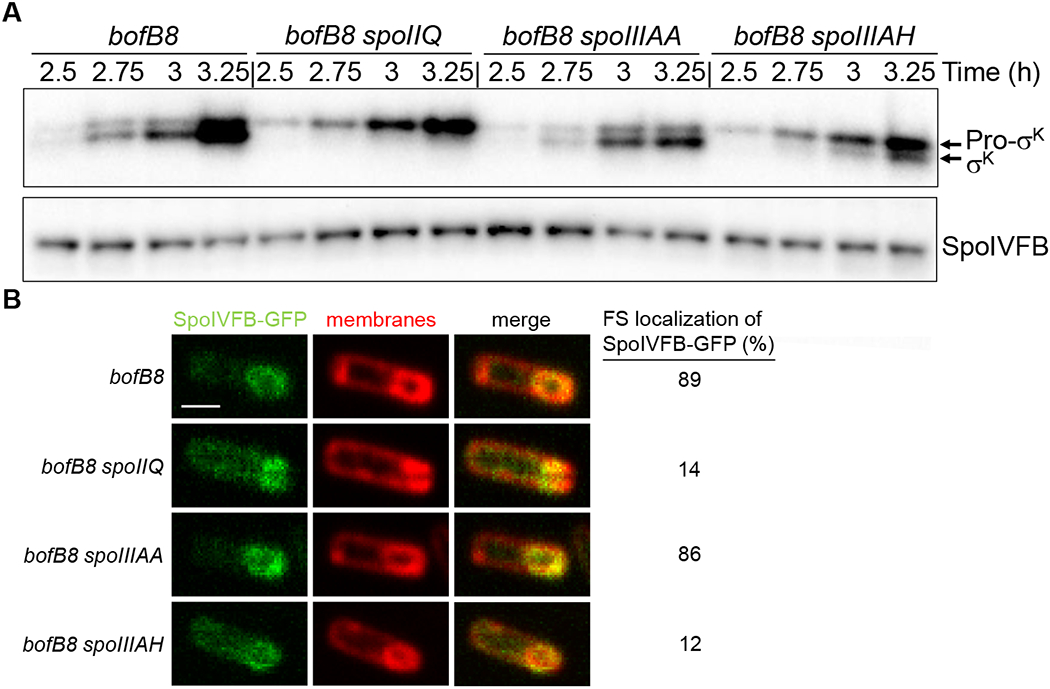
B. subtilis strains bearing the bofB8 mutation alone or in combination with a null mutation in spoIIQ, spoIIIAA, or spoIIIAH were starved to induce sporulation. All these strains have a null mutation in sigG, which is bypassed by the bofB8 mutation (Cutting et al., 1990). (A) Immunoblot analysis. Samples collected at the indicated times PS were subjected to immunoblot analysis using antibodies against Pro-σK and SpoIVFB. Representative immunoblots are shown. Similar results were observed for at least two biological replicates. (B) Confocal fluorescence microscopy. The native spoIVFB gene was C-terminally fused to gfp in the indicated strains, samples were collected at 3 h PS, and FM 4-64 was added to stain membranes. Images of fluorescence from SpoIVFB-GFP and membranes, and the merged images, are shown in the indicated columns, for representative sporangia with FS-localized (bofB8 and bofB8 spoIIIAA) or mislocalized (bofB8 spoIIQ and bofB8 spoIIIAH) SpoIVFB-GFP. In the top, left panel the scale bar = 1 μm. The percentage of sporangia (100-150 counted; rod-shaped cells were not counted) with FS-localized SpoIVFB-GFP is shown in the rightmost column.
