SUMMARY
Exposure to excessive sound causes noise-induced hearing loss through complex mechanisms and represents a common and unmet neurological condition. We investigate how noise insults affect the cochlea with proteomics and functional assays. Quantitative proteomics reveals that exposure to loud noise causes proteotoxicity. We identify and confirm hundreds of proteins that accumulate, including cytoskeletal proteins, and several nodes of the proteostasis network. Transcriptomic analysis reveals that a subset of the genes encoding these proteins also increases acutely after noise exposure, including numerous proteasome subunits. Global cochlear protein ubiquitylation levels build up after exposure to excess noise, and we map numerous posttranslationally modified lysines residues. Several collagen proteins decrease in abundance, and Col9a1 specifically localizes to pillar cell heads. After two weeks of recovery, the cochlea selectively elevates the abundance of the protein synthesis machinery. We report that overstimulation of the auditory system drives a robust cochlear proteotoxic stress response.
Graphical Abstract
In Brief
Jongkamonwiwat et al. perform quantitative proteomic analysis of mouse cochlear extracts immediately after noise exposure. They discover that many proteins have elevated levels and the proteostasis network is activated. During recovery, ribosomal proteins are upregulated. These results show that loud noise causing hearing loss results in cochlear proteotoxicity.
INTRODUCTION
Noise-induced hearing loss (NIHL) represents the primary cause of acquired hearing loss in the industrialized world (Carter et al., 2014; Saunders and Griest, 2009). Exposure to damaging levels of noise occurs through occupational, residential, and recreational activities. Sound pressure level (SPL) is measured in decibels (dB), the general unit used for measuring hearing and determining hearing thresholds. Decibels have a log10 scale, and a 20 dB sound is 10 times louder than a 10 dB sound. Sound pressure, frequency content, duration of sound exposure, and characteristics of the individual all play central roles in NIHL (Rabinowitz, 2000). For a sense of scale, Occupational Safety and Health Administration (OSHA) guidelines allow exposure to 85 dB for 8 h a day.
The cochlea is the part of the inner ear that converts mechanical vibrations into nerve impulses and transmits them to the brain. Inner hair cells (IHCs), outer hair cells (OHCs), and auditory nerve fibers (ANFs) are the specialized components within the organ of Corti (OC) that play central roles in facilitating hearing. Loud noise affects many elements of the cochlea, and in general, the most predominant substrates are stereocilia bundles composed of rigid cross-linked actin filaments, presynaptic hair cell (HC) ribbons, and ANF terminals (Liberman, 2017). Intense noise also reduces tectorial membrane calcium levels and cochlear blood flow (Le Prell et al., 2007; Strimbu et al., 2019), and it can cause HC death through numerous signaling pathways (Cheng et al., 2005). Intense noise at a SPL greater than 115 dB causes direct mechanical trauma to the fragile cells and tissues of the inner ear, physically impairing the transduction of auditory information (Henderson and Hamernik, 1995; Le Prell et al., 2007; Liberman and Dodds, 1984). Lower levels of injurious noise cause minimal direct physical damage to the cochlea but result in the loss of synaptic connections between IHCs and spiral ganglion neurons (SGNs), causing cochlear synaptopathy. Robust stimulation of the auditory system also imposes a high metabolic demand on many cochlear cell types (Hill et al., 2016), a condition that drives an increase in oxygen consumption, free radical production, and excitotoxicity, which also contribute to NIHL (Le Prell et al., 2007; Lu et al., 2014; Maulucci et al., 2014).
NIHL is categorized based on the period of postexposure hearing impairment (Liberman, 2016). Exposure to loud noise for hours (e.g., rock concerts or nightclubs) brings about an acute yet transient attenuation of hearing sensitivity, called a temporary threshold shift (TTS), which decreases auditory sensitivity for days to weeks. In contrast, more severe auditory insults causing a permanent threshold shift (PTS) result in partial or complete sensorineural hearing loss. In laboratory settings, threshold shifts that fail to recover to baseline levels 2 weeks following exposure are generally considered permanent in rodents (Kujawa and Liberman, 2006; Liberman, 2016; Ryan et al., 2016). Despite these clear physiological definitions, we lack a comprehensive understanding of the complex molecular mechanisms responsible for noise-induced threshold elevation and the processes responsible for hearing threshold recovery.
Loud noise triggers robust synaptic transmission between HCs and ANFs and consequently drives feedback inhibition through olivocochlear efferent nerve fibers (Kujawa and Liberman, 2009; Lin et al., 2011). ANF terminals are sensitive to insults and dramatically swell shortly after exposure to loud or very loud noise. In the process, they physically uncouple presynaptic HC ribbon membranes from postsynaptic ANF membranes (Shi et al., 2016). Interestingly, swelling is reversible and subsides within a few days of exposure (Robertson, 1983). However, after exposure to loud noise causing cochlear synaptopathy, up to half of IHC ribbon synapses are permanently lost (Kujawa and Liberman, 2009; Ruel et al., 2007). Noise overexposure can also trigger metabolic decompensation, resulting in the swelling of nuclei and mitochondria, cytoplasmic vesiculation (Kim et al., 2014), and apoptosis (Op de Beeck et al., 2011).
If we can obtain a deep understanding of the mechanisms contributing to NIHL, in particular cochlear synaptopathy, we may accelerate our ability to develop strategies for prevention and treatment. Mass spectrometry (MS)-based proteomics provides an opportunity to investigate complex biological phenomena by quantifying the abundance of thousands of proteins. To investigate how auditory overstimulation influences the cochlea, we quantified the cochlear proteome acutely after noise exposure across three intensity levels. Unexpectedly, we found hundreds of proteins with elevated fold changes (FCs) after noise exposure relative to controls. In particular, exposure to noise with increasing levels of intensity drives a dose-dependent elevation of many proteostasis factors, including nearly the entire proteasome and many protein chaperones. Independent proteomic and biochemical analysis confirmed that exposure to intense noise triggers a cochlear protein quality control response. Immunofluorescence (IF) and bioinformatics revealed that a panel of proteins altered after noise exposure are expressed by HCs, SGNs, and additional cochlear cells. RNA sequencing (RNA-seq) analysis revealed that a subset of genes encoding proteins with elevated FCs, including many proteasomal subunits, also have elevated FCs after exposure to loud or very loud noise. Proteome-wide analysis of cochlear protein ubiquitylation after exposure to loud or very loud noise revealed that many proteins become selectively modified on lysine residues. Finally, we measured the cochlear proteome after two weeks of recovery and found that nearly the entire ribosome had elevated FCs after exposure to loud noise, indicating that protein synthesis is a contributor to threshold recovery and suggesting that noise exposure causing hearing loss accelerates protein turnover.
RESULTS
Short-Term Noise Exposure Causing Temporary or Permanent Hearing Loss in Mice
We set out to advance our understanding of how auditory overstimulation causes cochlear synaptopathy by first using cochlear functional assays to determine the degree of temporary and permanent hearing loss across a panel of noise exposure intensities. Young adult FVB mice were exposed to band-pass-filtered white noise (6–18 kHz) for 30 min at three intensity levels: ambient (70 dB SPL), loud (94 or 100 dB SPL), or very loud (105 dB SPL). To deeply assess of how cochlear protein levels become altered by excess noise, we exposed both awake and anesthetized mice. Awake noise exposure is more physiological relevant, but anesthetized exposure provides quicker access to the cochlear tissue, because it can be extracted immediately. Auditory brainstem response (ABR) tone and click, as well as distortion product otoacoustic emission (DPOAE) hearing measurements, were performed before and 1, 7, and 14 days after noise exposure. Overall, the level of threshold shift was greater for anesthetized mice compared with awake mice. We found that both anesthetized and awake mice exposed to 65–70 dB SPL noise, representing ambient noise levels, resulted in only a slight increase in click and tone ABR thresholds 1 day after noise (Figure S1). Wave I amplitude, which indicates the strength of synaptic transmission primarily between IHCs and ANFs, in the absence of alterations to DPOAEs, was also unaffected after exposure to 70 dB SPL noise. Noise exposures at 94 dB SPL had similar recovery profiles after 7 or 14 days, and we observed a near-complete recovery of hearing thresholds after two weeks (Figure S1). Exposure to 100 dB SPL noise acutely elevated both ABR and DPOAE thresholds compared with 94 dB SPL. However, after 7 and 14 days of recovery, ABR thresholds recovered to a large degree, but not to baseline levels (Figure S1). Consistent with structural damage to the mechanotransduction machinery or ANFs, 100 dB SPL was the lowest exposure level found to permanently reduce wave I amplitudes. Noise exposure at 105 dB SPL caused significant elevations in threshold levels in both DPOAE and ABR analyses. Wave I amplitudes were also significantly reduced, with minimal recovery of amplitudes and thresholds after two weeks, indicative of permanent damage. Anesthetized mice had larger threshold shifts 1 day after noise compared with mice that were noise exposed while awake at 94, 100, and 105 dB SPL. However, after 7–14 days, their threshold levels recovered to a similar degree (Figure S1). In summary, 30-min exposures at 70 dB SPL caused negligible hearing impairments, 100 dB SPL exposures resulted in both TTS and PTS in anesthetized mice, awake exposure to 94 dB SPL resulted almost exclusively in TTS, and exposure to 105 dB SPL resulted predominantly in PTS in both anesthetized and awake mice.
Damaging Noise Exposure Progressively Remodels the Cochlear Proteome
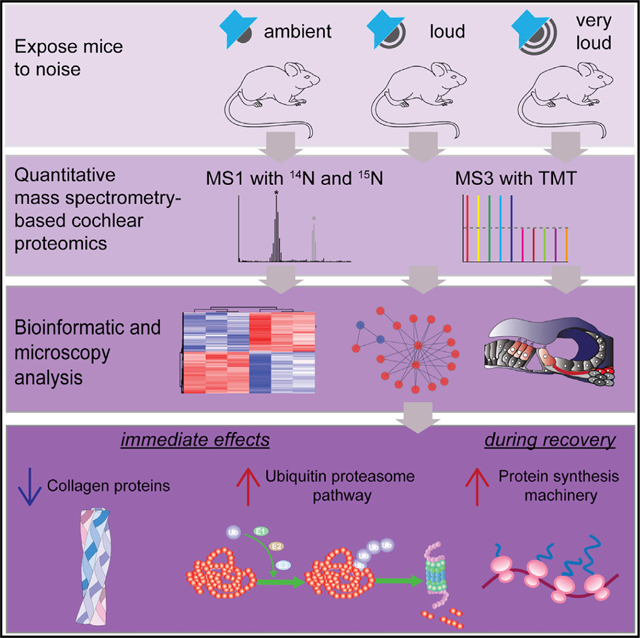
To investigate how exposure to loud and very loud noise affects the mouse cochlear proteome, we performed an initial quantitative proteomic screen with cochlear extracts from mice metabolically labeled with the heavy stable isotope 15N. We pooled cochleae from multiple 15N-labeled animals to facilitate relative quantification of unlabeled 14N proteins from experimental cochleae, serving as an internal standard for relative quantitative proteomic analysis (Butko et al., 2013). In this way, changes in protein because of noise exposure can be assessed by comparing the cochlear proteome of unlabeled noise-exposed 14N mice, which contains light proteins relative to unexposed cochleae containing heavy proteins. We mixed light and heavy cochlear extracts 1:1, digested the proteins to peptides, and performed liquid chromatograph with liquid chromatography-tandem mass spectrometry (LC-MS/MS)-based proteomic analysis (Link et al., 1999; Washburn et al., 2001). To control for potential quantification errors, we used a ratio-of-ratios analysis paradigm (MacCoss et al., 2003) and related each noise exposure condition relative to a group of mice subjected to the same stress of being placed in a sound chamber but without acoustic exposure (Figure 1A). Overall, we identified 738, 1,256, and 1,571 significantly altered proteins in cochleae exposed to ambient, loud, and very loud noise, respectively (Figure 1B; Table S1). Many proteins were altered in multiple exposure conditions, and 582 proteins were significantly altered by both loud and very loud noise. Next, we compared the levels of the significantly altered proteins across the three datasets. We found that the difference in average protein FC is significantly elevated in a noise-intensity-dependent manner (Figure 1C). To assess the quantitative variability among biological replicates, we examined the overall 14N/15N peptide ratios and performed regression analysis to determined threshold log2 fold differences (based on protein ratios) from the ambient noise exposure proteomic datasets with the lowest correlations. The normalized 14N/15N peptide ratio distributions were similar among biological replicates (Figure S2A), and the threshold levels of log2 fold cutoffs were determined at 1.24 and −1.35 for elevated and reduced levels, respectively (Figure S2B). To home in on the most confidently altered proteins, we used these thresholds to filter our data and again found many more proteins with elevated, rather than decreased, steady-state abundance. Specifically, we found 21, 115, and 226 proteins with significantly elevated levels in the ambient, loud, and very loud datasets, respectively (Figure 1D). To visualize the cochlear proteome datasets, we graphed our results using volcano plots (Figures 1E–1G). In total, our proteomic analysis provided relative quantitation for >2,800 proteins in each paradigm. This representation also revealed that exposure to increasingly intense levels of noise skews the cochlear protein abundance distribution such that more proteins have significantly elevated levels relative to levels measured in controls. A panel of proteins (e.g., Psmc5 and Uba2) had a noise-dose-dependent increase in their levels after auditory stimulation. A smaller number of proteins (e.g., Col9a1) had a noise-dose-dependent reduction in their levels relative to controls.
Figure 1. Noise Exposure Causing Hearing Loss Unbalances the Cochlear Proteome and Trigger a Proteostasis Stress Response.
(A) Analysis scheme to determine how noise exposure causing hearing loss acutely influences cochlear protein fold change (FC) using 15N-labeled cochlear extracts (gray) as internal standards and LC-MS/MS-based quantitative proteomics (n = 4–7 mice per group).
(B) Venn diagram of proteins with significantly altered FCs (B.H. p < 0.05) across all three levels of exposure. Values in parentheses represent the total number of significantly regulated proteins in each condition.
(C) Summary boxplots of proteins with significantly altered FCs. FCs were significantly elevated by exposure to increasingly intense noise (ambient = 0.175 ± 0.539, loud = 0.446 ± 0.627, and very loud = 0.572 ± 0.756, mean ± SD).
(D) Number of proteins with significantly altered abundance and meeting the threshold log fold differences(TLFD) threshold.
(E–G) Volcano plots depicting cochlear proteome remodeling after the indicated noise exposures for 30 min. Proteins that satisfied both the statistical cutoff (B.H. p < 0.05) and the TLFD threshold are in red (elevated) or blue (decreased).
(H and I) Enrichment analysis of significantly regulated proteins based on (H) the GO cellular component term and (I) the GO molecular function term.
(J) Number of proteins associated with the UPS and HSP chaperone networks are enhanced in a noise-dependent manner based on STRING-protein-protein interaction network functional enrichment analysis.
***p < 0.001 by Kruskal-Wallis (C); dotted line, False discovery rate (FDR) < 0.05 by Fisher’s exact test (H and I).
Proteasome Subunits and Protein Chaperones Have Selectively Elevated FCs after Exposure to Damaging Noise
To investigate the significantly regulated proteins, we performed Gene Ontology (GO) cell component (GO:CC) and GO molecular function (GO:MF) enrichment analysis with PANTHER (Thomas et al., 2006). Proteins altered exclusively in the very loud (105 dB SPL) dataset are significantly enriched for the GO:CC terms neuronal projections, synapses, and cell junctions, among other structures (Figure 1H), supporting previous evidence that excess noise alters cell junctions and synapses (Kujawa and Liberman, 2009). Proteins significantly altered in both loud and very loud datasets were significantly enriched for the terms cytoskeleton, cell projection, endopeptidase, and proteasome. Every significantly enriched GO:CC term in the loud dataset was also enriched in the very loud dataset. This finding provides support to the reproducibility of our analysis, because protein networks engaged acutely during loud noise exposure are expected to also be activated by very loud noise. Interestingly, all significantly enriched GO:MF terms are involved with binding, suggesting that noise exposure even at loud levels preferentially affects proteins known to assemble into higher-ordered structures (Figure 1I).
To mine our datasets further, we examined individual proteins significantly altered in multiple datasets. Overall, many proteins had progressively elevated abundance after exposure to increasingly intense noise. We identified 20 proteasomal proteins, 35 proteins associated with proteolysis, and 19 protein chaperones significantly enriched in the loud and very loud datasets, but not the ambient dataset (Figure S2C). To investigate the possibility that the significantly altered proteins are physically interact, we subjected the datasets to STRING analysis. We uncovered several protein-protein interaction networks that became increasingly populated across noise exposure level (very loud > loud > ambient). Most identified protein interaction hubs function in protein quality control, indicating that noise exposure triggers a proteotoxic response (Figure 1J).
Confirmation that Noise Exposure Unbalances the Cochlear Proteome
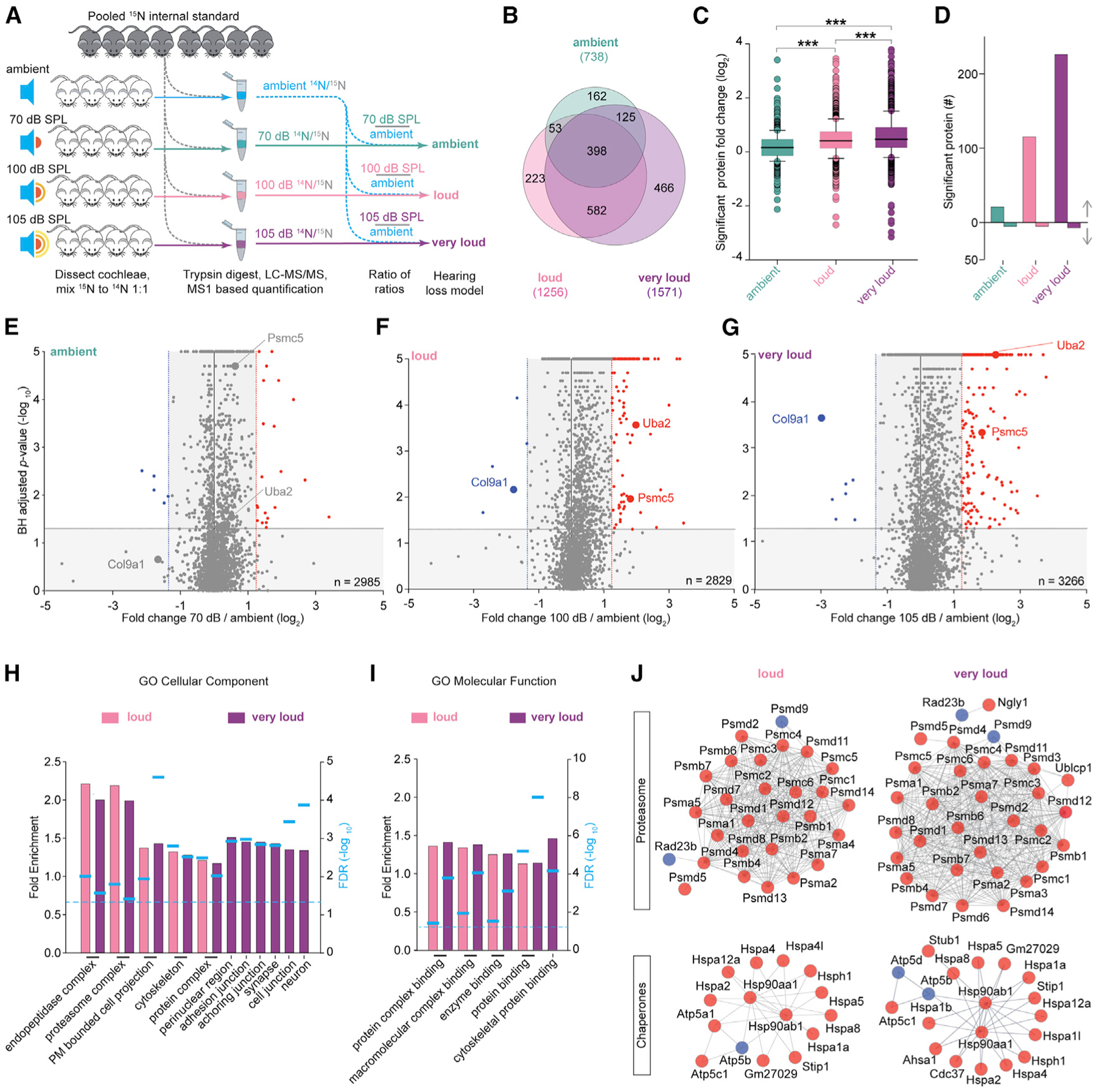
Western blot analysis for a few prominent proteins from the initial proteomic datasets verified that the actin cytoskeleton regulator Arpc2 and the proteasome ATPase subunit Psmc5 have elevated levels after 30 min of very loud noise exposure (Figure S3A). To assess changes in cochlear protein abundance by exposure to loud or very loud noise relative to ambient noise with a large-scale strategy, we performed two 10-plex tandem mass tag (TMT) experiments (He et al., 2019) (Figure S3B). In each experiment, we exposed three mice to loud noise (94 dB SPL), three mice to very loud noise (105 dB SPL), and four mice to ambient noise (65–70 or 70 dB SPL). The overall TMT channel peak intensities were similar across 10 channels in both experiments, showing that the chemical labeling was efficient and reproducible (Figure S3C). To assess the reliability of the TMT data, we assessed the protein abundance among the biological replicates and found that in general, they were well correlated (Figure S3D).
TMT analysis confirmed many protein abundance trends observed in the 15N-based quantitative proteomic analysis. In both TMT experiments, we quantified more than 3,500 proteins. The relative protein abundance distribution was, on average, significantly elevated in mice cochleae exposed to loud versus ambient noise (Figure S3E). Furthermore, the global FC (i.e., loud or very loud noise relative to the ambient controls) of the significantly altered proteins was skewed toward proteins with higher ratios (Figure S3F; Table S2). Numerous cytoskeletal, heat shock, and proteasome protein subunits were confirmed to have elevated FCs in a noise-intensity-dependent manner, and again, we found multiple collagen proteins to be reduced (Figures 2A and S3G). In total, 440 or 147 proteins were found to be significantly altered in TMT and 15N analyses after exposure to loud or very loud noise relative to ambient intensity, respectively (Table S2). We plotted the relative protein FCs for the 147 proteins found to be significantly altered in both TMT and 15N analyses (Figure 2B). Of the proteins found to be significantly altered by both quantitative proteomic strategies, 75.0% proteins with elevated levels and 66.6% proteins with reduced levels were found with the same FC trends in both TMT and 15N analyses (Figure 2C). We went on to quantify the absolute abundance of the actin binding protein Tpm1 and the fibrillary protein Col1a1 with chemically synthesized heavy peptides and LC-MS/MS (Li et al., 2015). These experiments provided quantitative support of the 15N results, because they confirmed elevated and reduced levels, respectively (Figure 2D).
Figure 2. Confirmation that Noise Exposure Causing Hearing Loss Unbalances the Cochlear Proteome.
(A) Selected abundance plots for cytoskeletal, heat shock, proteasome, autophagy, synaptic, and collagen proteins with significantly elevated or reduced levels across noise exposures.
(B) Comparison of individual proteins with significantly altered FCs in very loud 15N and TMT datasets.
(C) Summary of protein FC trends for proteins significantly altered in both 15N and TMT very loud datasets. Of the 140 proteins found to be significantly altered by 15N, 66.6% of proteins with reduced levels and 75.0% of proteins with elevated levels were verified in TMT.
(D) Absolute quantification of Tpm1 (IQLVEEELDR) and Col1a1 (ALLLQGSNEIELR) based on the ratio of light and heavy (Arg+10) peptide-reconstructed MS1 chromatograms. The absolute abundance of Tpm1 was significantly elevated between loud versus ambient noise and very loud versus ambient noise (ambient = 0.233 ± 0.011, loud = 0.834 ± 0.0417, and very loud = 0.499 ± 0.0249, mean ± SD). The absolute abundance of Col1a1 was significantly reduced between loud versus ambient noise and very loud versus ambient noise (ambient = 0.428 ± 0.0214, loud = 0.215 ± 0.0107, and very loud = 0.114 ± 0.00570, mean ± SD, nanomoles per cochlea). n = 3 mice per noise exposure condition.
*p < 0.05, **p < 0.01, ***p < 0.001 by one-way ANOVA with Bonferroni correction (A); *p < 0.05, **p < 0.01 by one-way ANOVA with Holm-Sidak (D).
To investigate noise-induced cochlear proteome remodeling on a shorter time frame, we exposed mice to ambient, loud, or very loud noise (70, 100, and 105 dB SPL) for 15 min. ABR tone analysis revealed that the threshold shift was significantly elevated 1 day after noise exposure for mice exposed to 100 or 105 dB SPL, but not 70 dB SPL, compared with 7 or 14 days of recovery (Figures S4A–S4C). Mice in the 100 or 105 dB SPL 15-min exposure groups displayed robust recovery within 7 days; these exposures generated a nearly pure TTS. We again used the 15N cochlear ratio-of-ratios strategy to obtain relative measures of protein FC. We quantified more than 2,500 proteins in each paradigm, and similar to the 30-min exposure analysis, we found many more proteins with significantly elevated FCs compared with reduced FCs (Figures S4D–S4F; Table S3). In contrast to our findings with the 30-min exposures, we did not observe a noise-intensity-dependent increase in the number of significantly altered proteins after 15-min exposure (Figure S4G). However, when we directly compared protein abundance measured in both the 15-min and the 30-min datasets, we found that most proteins had the same trends (Figures S4H–S4M). This analysis confirmed 97, 135, and 164 proteins with reduced abundance and 204, 456, and 594 proteins with elevated levels after 70, 100, and 105 dB SPL exposure, respectively.
To assess how the cochlear proteome specifically responds to loud versus very loud noise (94 or 105 dB SPL), we carried out a 10-plex TMT-based quantitative proteomic analysis (Figure S5A). We quantified more than 2,300 proteins and found more significant proteins with relatively elevated FCs compared with reduced FCs after exposure to very loud versus loud noise (i.e., proteins with FC > 1 compared with proteins with FC < 1 [very loud/loud]) (Figure S5B). Among the proteins with the most significant FC <1 (very loud/loud) were Ckm, Tnni2, Tnnc2, and Myh8, which play key roles in acute fluctuating energy transduction and force regulation (Figure S5C; Table S4). Pvalb, a calcium-binding albumin protein localizing to ANF terminals, also had a significant FC <1 (very loud/loud) (Kujawa and Liberman, 2009). Of particular interest, numerous heat shock chaperones and several proteasome subunits had significant FC >1 (very loud/loud) (Figure S5D). This finding provides independent confirmation that the cochlear proteostasis network is engaged in a noise-intensity-dependent manner.
Reduced Ribbon Synapse Density, Modest Damage to the Mechanotransduction Machinery, and No Cell Death after Exposure to Loud Noise
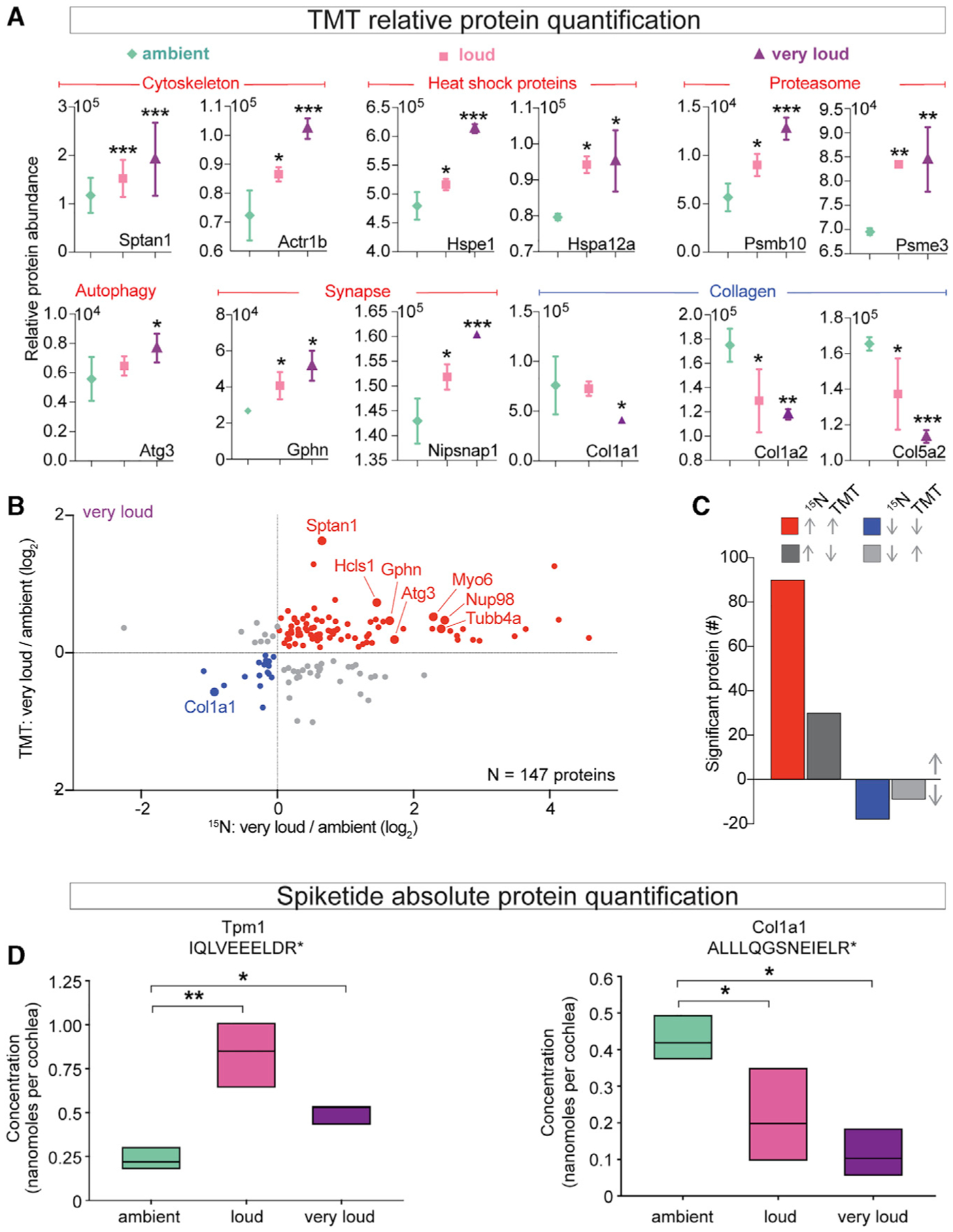
In the next set of experiments, we assessed potential damage to the mechanotransduction machinery, changes in ribbon synapse density, and changes in HC density across our 30-min noise exposure conditions. First, we used scanning electron microscopy (SEM) to assess physical damage to the OHC stereocilia after exposure to noise (Figure 3A). In the apical, middle, and basal regions of cochleae, stereocilia bundles generally appeared normal in mice exposed to ambient noise. However, we observed significantly more frayed OHC stereocilia bundles in the middle region from cochleae exposed to very loud levels compared with loud or ambient levels of noise (Figure 3B). The base was even more vulnerable, and we observed significantly more damaged stereocilia bundles after either loud or very loud noise compared with ambient noise (Figure 3B). Overall, the SEM-based characterization of stereocilia bundle damage was consistent with our DPOAE results (Figures S1G, S1K, and S1S).
Figure 3. Exposure to Loud and Very Loud Noise Alters Stereocilia Morphology and Reduces Synaptic Density across the Middle and Base of the Cochlea.
(A) Representative SEM images of stereocilia bundles from cochlea whole mounts prepared immediately after noise exposure from apical (8–12 kHz), middle (12–16 kHz), and basal (28–32 kHz) regions (i.e., 80%, 65%, and 40% of the distance from the base, respectively).
(B) Percentage of stereocilia bundles with visible fraying was significantly elevated following exposure to increasingly intense noise across the middle region (ambient = 7.48 ± 4.84, loud = 10.70 ± 5.59, and very loud = 49.35 ± 8.21, mean ± SD) and basal region (ambient = 3.54 ± 3.35, loud = 15.95 ± 5.26, and very loud = 26.48 ± 5.03, mean ± SD). The apex was largely unaffected by the intensity of the exposure (70 dB = 6.76 ± 4.64, 94 dB = 7.31 ± 4.78, and 105 dB = 9.58 ± 4.40, mean ± SD). n = 645, 585, and 719 bundles analyzed for ambient, loud, or very loud noise, respectively.
(C–H) Representative images and quantification of synapse density from apical, middle, and basal regions. Cochlear synaptic density within the apical region was significantly reduced in the very loud versus ambient groups (ambient = 14.20 ± 0.37, loud = 13.45 ± 1.04, and very loud = 12.00 ± 2.44, mean ± SD). Cochlear synapse density was significantly reduced in the middle and basal regions following exposure to both loud and very loud noise compared with ambient noise (middle: ambient = 17.52 ± 0.93, loud = 9.71 ± 1.90, and very loud = 9.18 ± 1.82; base: ambient = 13.97 ± 0.44, loud = ± 0.74, very loud = ± 1.98, mean ± SD).
(I) Representative images of HC density across the cochlea.
(J) Base had slightly reduced HC density in the very loud group compared with the loud and ambient groups (ambient = 55.19 ± 1.22, loud = 55.70 ± 1.01, and very loud = 50.87 ± 2.94, mean ± SD).
Scale bars, 5 μm (A), 10 μm (B–D), and 20 μm (C, E, and G). *p = 0.05, ***p < 0.001 by one-way ANOVA with Bonferroni correction (B, D, F, H, and J). n = 3 mice (A and B), 4 mice (C–H), and 3–6 mice (I and J).
Next, we analyzed cochlear ribbon synapse density after noise exposure (Figures 3C–3H). In the apical region (8–12 kHz) of the cochlea, we found that ribbon synapse density was significantly reduced after exposure to very loud noise, but not loud noise, compared with ambient controls. Synapse density in the middle region of the cochlea (12–16 kHz) was significantly reduced after exposure to loud or very loud noise compared with ambient controls. The base of the cochlea (24–28 kHz) was also found to have significantly reduced synapse density after exposure to loud or very loud noise compared with ambient controls.
To investigate the possibility that our noise exposure conditions cause a reduction of HC density, presumably by causing cell death, we performed IF analysis across the cochlea 14 days after exposure to ambient, loud, and very loud noise (Figure 3I). Consistent with previous findings in other mouse strains, we did not observe substantial HC death in our noise exposure conditions. We only detected a slight yet significant reduction in HC density per 100 μm after exposure to very loud noise, suggestive of increased HC death, in the base of the cochlea (Figure 3J).
HCs, SGNs, and Additional Cochlear Cell Types Express Proteins with Altered FCs after Noise Exposure
To begin to determine which cells express our candidate proteins, we extracted transcriptomic data from the gene expression for auditory research (gEAR) portal database (Hertzano and Elkon, 2012). We used transcriptomic datasets obtained from four defined cell populations (i.e., HCs and supporting cells: neuron, vascular endothelia, and mesenchyme) isolated by flow cytometry (Hertzano et al., 2011). As expected, we found that most noise-sensitive proteins were expressed by multiple cochlear cell types (Figures S6A–S6D). However, in this analysis, we identified a small number of our candidate proteins as predominantly expressed in specific cell types. For example, Hsp90b1, Hsp5, and Col9a1 are expressed predominantly by HCs or supporting cells. Snap25 is exclusively expressed by neurons, whereas Itga6, the laminin receptor, and Myh9, which is genetically linked to progressive sensorineural hearing loss, are expressed highly in the vascular endothelia (Canzi et al., 2016).
To identify proteins enriched in HCs, we used our previous proteomic analysis of GFP-expressing HCs isolated with fluorescence-activated cell sorting as a reference (Hickox et al., 2017) (Figure S6E). We compared the proteins enriched in HCs with those found to have significantly altered FCs after exposure to loud or very loud noise from 15N- or TMT-based quantitative proteomics, or vice versa. Interestingly, we identified 84 and 109 HC-enriched proteins with significantly altered FCs after exposure to damaging noise in our 15N and TMT experiments, respectively (Figure S6F, top). This indicates that a modest proportion (17.0% and 22.1%, respectively) of HC-enriched proteins was found with significantly altered levels in our proteomic analyses of cochleae exposed to damaging levels of noise. However, this population of HC-enriched proteins represents only a small fraction of the total proteins we found with significantly altered levels in cochlear extracts (3.5% and 4.6% for 15N and TMT, respectively) (Figure S6F, bottom).
Next, we exposed mice to ambient, loud, or very loud noise for 30 min and performed IF to investigate the cochlear protein localization patterns of selected protein candidates (Figure 4A). First, we performed IF analysis of OC whole mounts and midmodiolar sections with Col9a1 antibody. We observed a prominent signal in the region between IHCs and OHCs near the top of the tunnel of Corti, likely in the heads of the pillar cells, which was less prominent after exposure to loud or very loud noise compared with ambient noise (Figures 4B and S6G). We examined Hsp90b1 localization patterns, a molecular chaperone that processes secreted proteins and functions in the degradation pathway associated with the endoplasmic reticulum. Hsp90b1 levels are elevated in IHCs and OHCs after exposure to loud or very loud noise compared with ambient controls (Figure 4C). We also found that levels of the proteasome subunit Psmc5 are increased in IHCs after loud and very loud noise exposure (Figure 4D). To investigate proteostasis factors, we examined VCP, which is a part of the ubiquitin-proteasome system (UPS). We found that VCP levels are elevated in a noise-intensity-dependent manner consistent with the 15N analysis in IHCs, OHCs, and SGNs (Figures 4E, S6H, and S6I). Finally, the actin binding proteins Arpc2 and Uba2 (E1 for SUMO1–SUMO4) were found be progressively elevated in SGNs after noise exposure, whereas latent-transforming growth factor b-binding protein 4 (Ltbp4) was reduced (Figure 4F).
Figure 4. Proteins Regulated during Noise Exposure Are Expressed by HCs, SGNs, and Supporting Cells within the Cochlea.
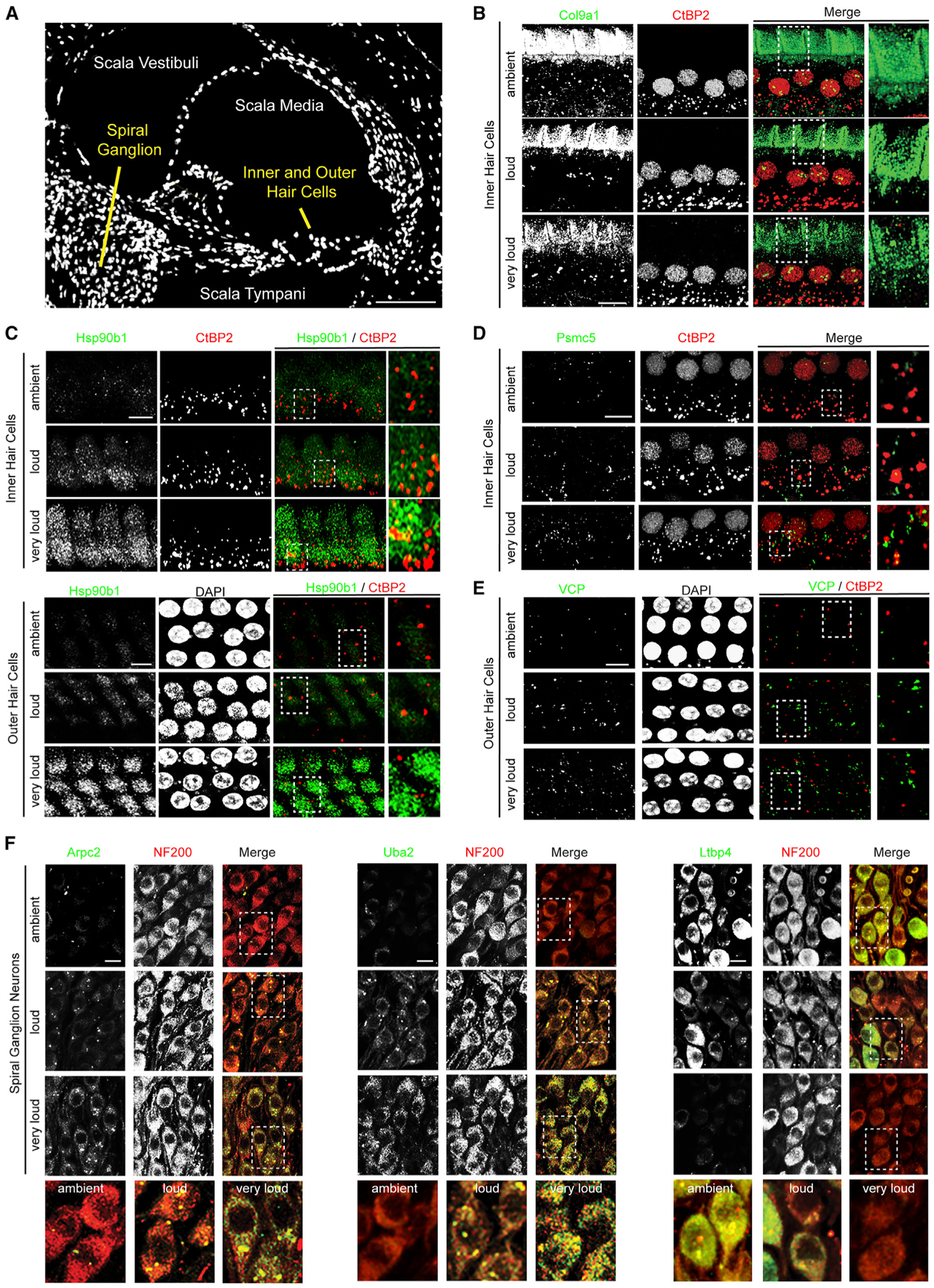
(A) DAPI-stained midmodiolar cochlear section.
(B) Col9a1 signal decreases in the area above the tunnel of Corti following noise exposure. Green, Col9a1; red, CtBP2.
(C) Hsp90b1 levels are elevated in both IHCs and OHCs after exposure to increasing intensities of noise. Green, Hsp90b1; red, CtBP2; white, DAPI.
(D) Psmc5 levels increase after exposure to increasing noise intensity within IHCs. Green, Psmc5; red, CtBP2.
(E) Vcp levels are elevated in the OHCs after exposure to increasingly damaging levels of noise. Green, Vcp; red, CtBP2; white, DAPI.
(F) SGNs from the second ganglion bundle from the top of the cochlea (12–16 kHz) in midmodiolar sections. Left: Arpc2 levels are elevated in SGNs after exposure to increasingly damaging levels of noise. Middle: Uba2 levels progressively increase within SGNs with increasing noise exposure. Right: LtBP4 levels are reduced in SGNs after exposure to increasingly damaging levels of noise. Green, Arpc2, Uba2, and LtBP4 (left, middle, and right); red, NF200.
Scale bars, 100 μm (A) and 10 μm (B–F). Representative images from n = 2 animals.
Noise Exposure Causing Hearing Loss Drives Cochlear Gene Expression
To investigate whether elevated protein levels can be explained by increased gene expression, mice were exposed to ambient, loud, or very loud noise for 30 min and cochlear extracts were analyzed with RNA-seq analysis (Li and Dewey, 2011). Visualization of global gene expression with violin plots showed similar overall distributions across the datasets (Figure 5A). To investigate the reproducibility of our transcriptomic analysis, we assessed correlation of gene expression patterns across our datasets. Biological replicates clustered by noise exposure intensity and gene expression patterns from cochleae exposed to very loud noise were more similar to ambient controls than to cochleae exposed to loud noise (Figure 5B). We also performed principal-component analysis (PCA) and found that the biological replicates clustered and datasets from mice exposure to very loud noise were more similar to those of mice exposed to ambient rather than loud noise (Figure 5C).
Figure 5. RNA-Seq Analysis Demonstrated an Overall Increase in Cochlear Gene Expression following Exposure to Loud or Very Loud Noise Compared with Ambient Levels.
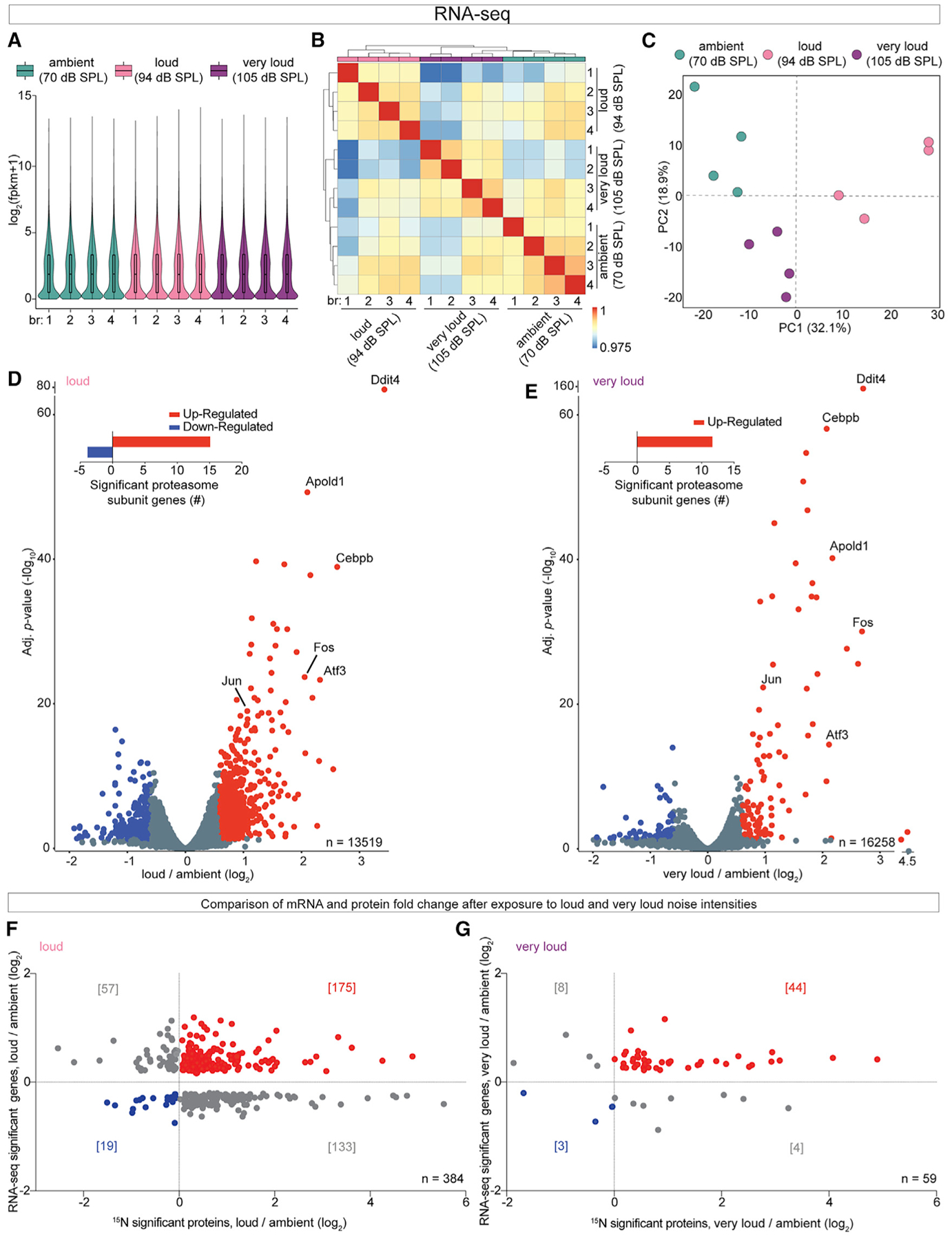
(A) Global gene expression patterns with violin plots showed similar overall distributions across the datasets.
(B) Correlation of the gene expression matrix demonstrated that biological replicates cluster based on noise exposure intensity. Cochleae exposed to very loud noise were more similar to ambient controls rather than to cochleae exposed to loud noise.
(C) PCA shows the biological replicates cluster and demonstrates that the very loud noise condition is more similar to the ambient condition than the loud noise condition based on transcriptomics.
(D and E) Noise-induced changes in cochlear gene expression graphed as FCs relative to ambient controls. Nearly twice as many genes with significantly altered FCs were found altered in the loud compared with the very loud datasets. Insert: 15 and 12 genes encoding individual proteasome subunits had significantly elevated FCs in loud and very loud datasets, respectively. Genes meeting the statistical cutoff (B.H. p < 0.05) are in red (elevated) or blue (decreased).
(F and G) Comparison of corresponding mRNA and protein FCs after exposure to loud (F) or very loud (G) noise relative to ambient controls. Roughly 45.6% and 74.6% of proteins with elevated FCs also had elevated mRNA abundance in the loud and very loud datasets, respectively. n = 4 mice per group (A–G).
Noise-induced changes in cochlear gene expression were graphed as FCs after exposure to loud or very loud noise relative to the ambient controls on volcano plots (Figures 5D and 5E; Table S5). We found nearly twice as many genes with significantly altered FCs in the loud dataset compared with the very loud dataset. In both analysis paradigms, there were slightly more genes with elevated FCs rather than reduced FCs. Interestingly, 15 and 12 genes encoding individual proteasome subunits had significantly elevated FCs in loud and very loud datasets, respectively, suggesting elevated levels of proteasome proteins observed in the proteomics at least partially result from elevated gene expression in response to noise exposure. However, none of proteasome subunit transcripts were among the genes with the most robustly elevated FCs. Ddit4, Atf3, Fos, Jun, and Cebpb were among the genes with the most dramatically elevated gene expression FCs after exposure to either loud or very loud noise. Ddit4 is rapidly induced after various conditions (Kimball and Jefferson, 2012). Elevated Ddit4, Atf3, Fos, Jun, and Cebpb gene expression has been observed in several transcriptomic studies of hearing loss (Low et al., 2010; Schiavon et al., 2018). Apold1, an endothelial early response gene that regulates endothelial cell signaling, was also robustly upregulated (Regard et al., 2004).
We compared mRNA and protein FCs after exposure to loud or very loud noise relative to ambient controls. Overall, there were 384 and 59 mRNA protein pairs identified with significantly altered levels in the loud and very loud datasets, respectively (Figures 5F and 5G; Table S5). We found that 45.6% or 74.6% of proteins with elevated FCs also had elevated mRNA abundance in the loud and very loud datasets, respectively, suggesting that increased gene expression contributes to, but is not solely responsible for, elevated protein FCs observed after exposure to damaging levels of noise.
UPS Is Activated after Exposure to Loud and Very Loud Noise
To explore the possibility that excess noise activates the cochlear proteostasis network, we investigate protein ubiquitylation with three complementary assays. Mice were exposed to ambient, loud, or very loud noise; cochlear extracts were prepared; and ubiquitylated proteins were biochemically enriched with tandem ubiquitin binding entities (TUBEs) (Hjerpe et al., 2009) (Figure 6A). Western blot analysis of the TUBE-purified material suggested that exposure to loud or very loud noise compared with ambient noise causes an increase in protein ubiquitylation (Figure 6B). Next, to assess the identity of the presumably ubiquitylated proteins, we repeated the experiment with three independent groups of mice and analyzed the material enriched with TUBEs by LC-MS/MS-based proteomics. We identified 328 proteins that were enriched or exclusive identified in at least two purifications from cochlear extracts exposed to loud or very loud noise compared with ambient controls. In addition, 108 of these proteins were found to have elevated protein FCs in the 15N experiments (Figure 6C).
Figure 6. The Degree of Ubiquitylation within the Cochlea Rises at Progressively Increasing Levels of Noise Exposure Compared with Ambient Controls.
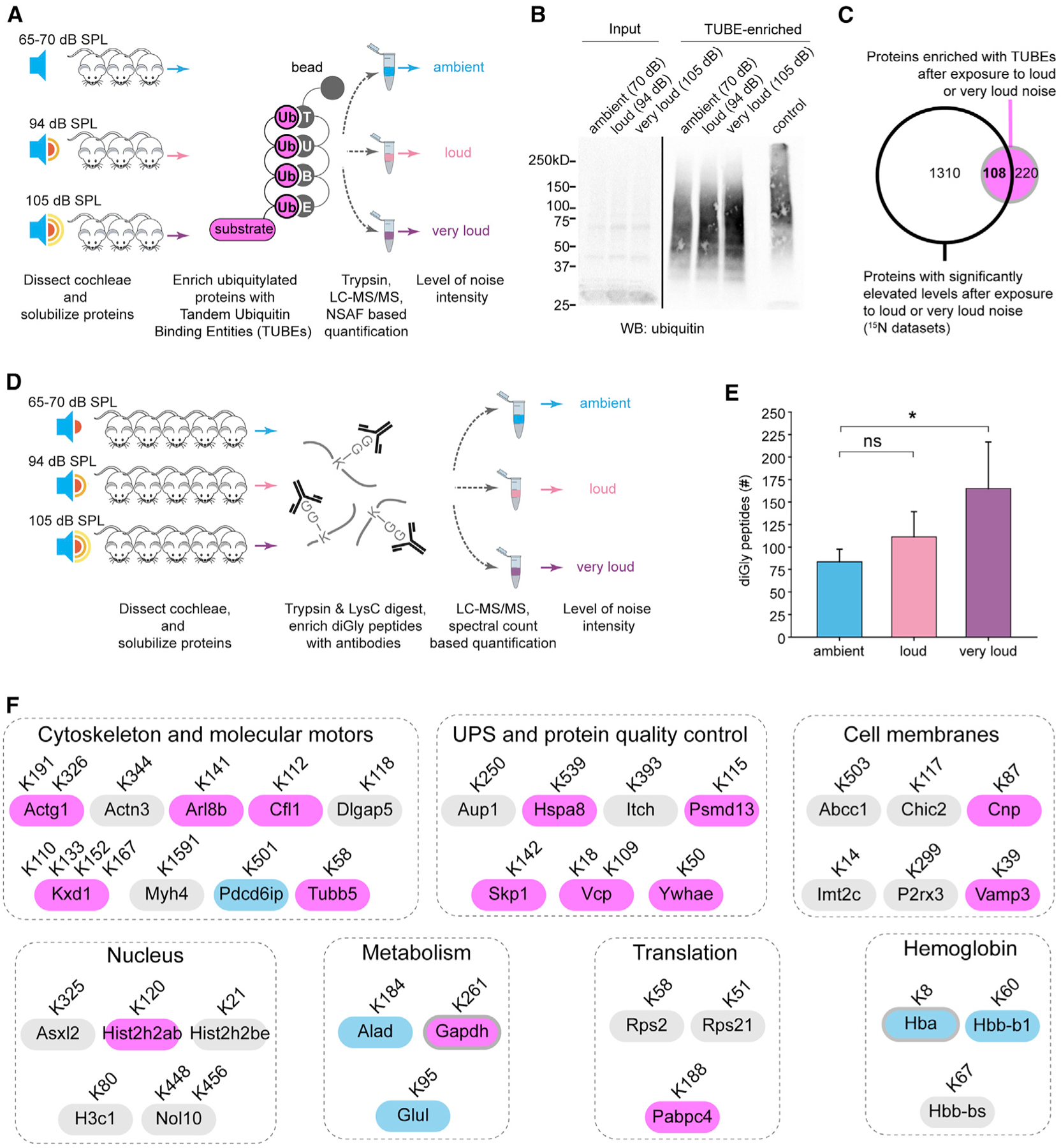
(A) Analysis scheme to identify ubiquitylated proteins with TUBEs.
(B) Western blot analysis of the TUBE-purified material, suggesting that exposure to loud or intense noise compared with ambient noise increases global protein ubiquitylation. Control, MG132-treated HEK293 lysate.
(C) TUBE-purified material from each noise exposure group (n = 4 mice, repeated 3 times) analyzed by LC-MS/MS. In total, 328 proteins were enriched (based on normalized spectral abundance factor (NSAF)) or exclusively identified in at least two purifications from either the loud or the very loud group compared with ambient controls (pink). Comparison of proteins found with elevated protein FCs in 15N experiments showed 108 proteins that were also enriched based on TUBE assay with increasing noise exposure intensities.
(D) Experimental schematic for the diGly LC-MS/MS assay to identify peptides with C-terminal Arg-Gly-Gly ubiquitin residues.
(E) In total, 324 diGly-containing peptides from 202 proteins were identified. Significantly more diGly peptides were identified in cochlear extracts from mice exposed to very loud noise compared with ambient noise (165.0 + 51.7 compared with 83.7 + 14.2 mean ± SD; p < 0.05).
(F) 46 diGly peptides from 36 proteins were identified in cochlear extracts from at least two biological replicates exposed to loud or very loud noise but were absent from extracts from ambient-noise-exposed mice. 15 of these proteins were found with elevated FCs, including Actg1, Tubb5, Kxd1, Arl8b, Vcp, and Vamp3 (purple), whereas only 5 had reduced FCs (blue) in the 15N experiments.
*p < 0.05 by Kruskal-Wallis and Tukey pairwise multiple comparison (E).
To directly examine noise-induced cochlear protein ubiquitylation, we repeated our noise exposures in additional cohorts of mice and purified the ubiquitylated peptides with antibodies for the diGly remnant due to cleavage of the C-terminal Arg-Gly-Gly sequence of ubiquitin (Peng et al., 2003b; Xu et al., 2010) (Figure 6D). We identified 324 diGly peptides from 202 proteins in our analysis of cochlear extracts. In support of elevated cochlear protein ubiquitylation due to excess noise, we identified significantly more diGly peptides in cochlear extracts from mice exposed to very loud noise compared with ambient noise (Figure 6E). Interestingly, 46 diGly-containing peptides from 36 proteins were identified in cochlear extracts from at least two biological replicates exposed to loud or very loud noise but were below the limit of detection in cochlear extracts from ambient noise exposure. These proteins predominantly associate with the cytoskeleton, UPS, membranes, the metabolism, and the nucleus. Fifteen of these proteins had elevated FCs in our 15N-based analysis, including Actg1, Tubb5, Kxd1, Arl8b, Vcp, and Vamp3, whereas only five had reduced FCs in the 15N experiments (Figure 6F; Table S6).
Protein Translation Machinery Has Selectively Elevated FCs during the Recovery Period
To identify molecular mechanisms underlying the recovery of hearing sensitivity, we analyzed the cochlear proteome after a two-week recovery period with two experimental designs. First, we exposed a cohort of mice to either loud or very loud noise and quantified the cochlear proteome acutely or after two weeks of recovery with TMT-multiplex quantitative proteomics (Figure 7A). The hypothesis is that proteins with elevated FCs (i.e., recovery versus acute) after exposure to loud levels of noise serve as beacons for pathways contributing to hearing sensitivity recovery. Based on our ABR analysis, mice exposed to very loud noise may slightly recover sensitivity in their threshold for hearing but predominantly represent models of permanent hearing loss. Overall, we quantified 4,626 proteins and found more proteins with elevated rather than reduced FCs (recovery/acute) after both loud and very loud noise exposure (Figures 7B and 7C; Table S7). To assess these datasets, we again performed GO:MF, GO:CC, and GO biological process (GO:BP) enrichment analysis of the significantly altered proteins. To our surprise, we found that the panel of proteins significantly altered in the very loud datasets was not enriched in the ontologies, whereas the loud noise datasets were significantly enriched with several ontologies (Figure 7D). The GO:MF analysis revealed that proteins associated with protein synthesis are selectively elevated during the recovery period after exposure to loud noise. Specifically, the ontologies significantly enriched based on GO:CC and GO:BP are associated with ribosomes or ribonucleoproteins and translation or metabolism, respectively. To visualize the protein FCs of the significantly altered proteins during recovery, we generated heatmaps, and consistent with the GO analysis, many ribosomal proteins co-clustered (Figure 7E). To investigate the molecular basis of hearing sensitivity recovery, we performed another TMT experiment but compared protein FCs after two weeks of exposure to very loud or loud noise with protein FCs exposed to ambient noise. Again, we found evidence that the translational machinery has elevated levels during recovery, but in this paradigm, we observed this phenomenon in both the loud versus ambient datasets and the very loud versus ambient datasets (Figure S7). Altogether, the protein translation machinery has elevated levels two weeks after noise exposure, which suggests that protein synthesis may play a key role in the recovery phase of noise exposure.
Figure 7. Proteomic Assessment of the Molecular Mechanisms Underlying the Recovery of Hearing Sensitivity.
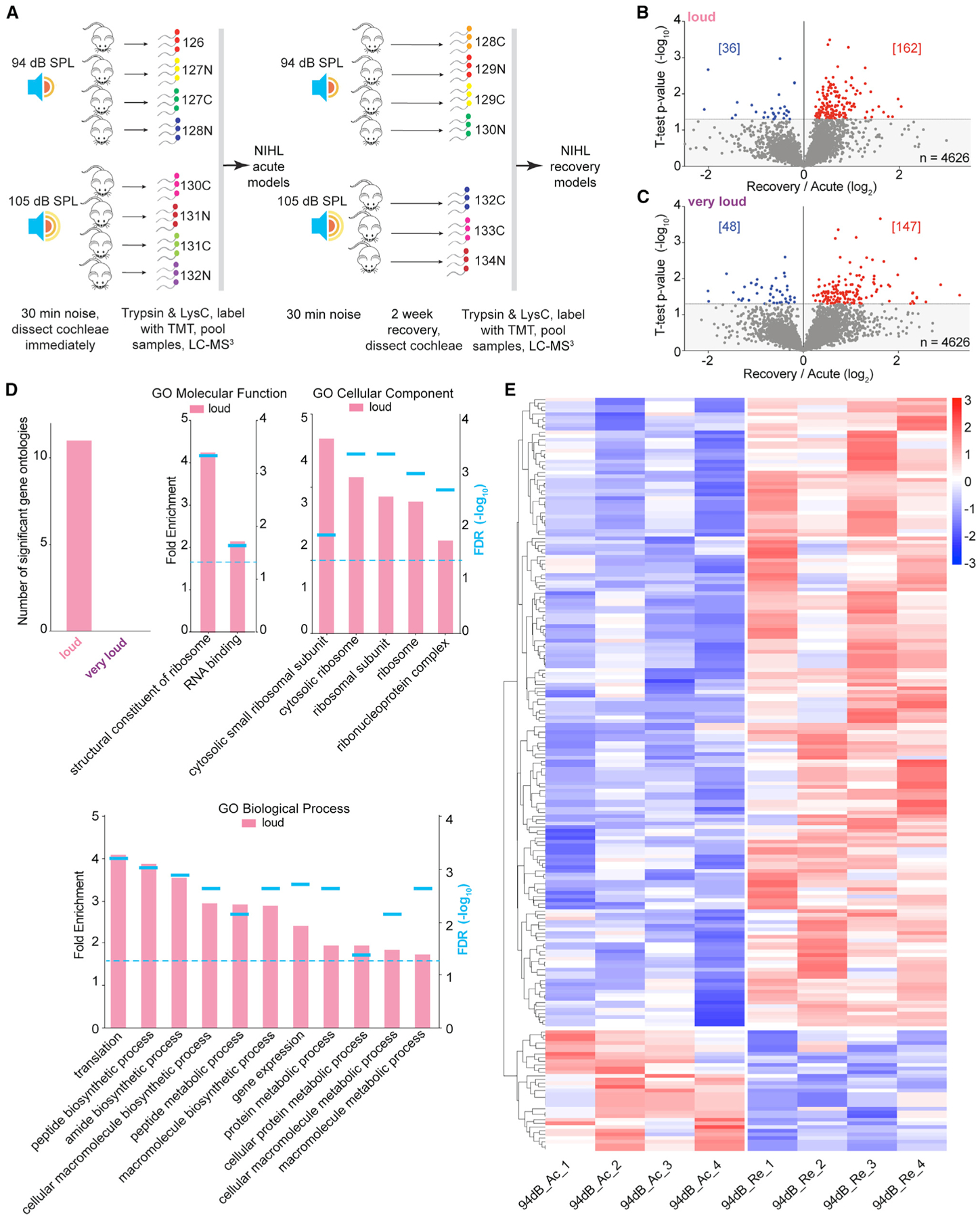
(A) Quantitative proteomic analysis workflow with 15-plex TMT to determine molecular processes associated with the recovery phases of loud and very loud noise exposure (n = 3–4 mice).
(B and C) Volcano plot of the cochlear proteome comparing the acute and recovery phases within loud (B) or very loud (C) noise for 30 min. Proteins meeting the statistical cutoff (t test, p < 0.05) are shown in either red (elevated) or blue (reduced).
(D) Number of significantly enriched GO:BP categories in the loud and very loud datasets (top left). Summary of GO enrichment analysis of significantly regulated proteins in the loud noise exposure condition. Dotted line, FDR < 0.05 by Fisher’s exact test.
(E) Heatmaps depicting protein FCs for the significant proteins in the acute phase versus the recovery phase. Ac, acute; Re, recovery; n = 391 proteins.
DISCUSSION
Our proteomic results add to the already compelling literature highlighting the inherently high complexity of the cochlear response to noise exposure. For example, we found that cochlear overstimulation causes cochlear structural alterations, changes in gene expression, elevated protein levels, and increased levels of global protein ubiquitylation. We focused on noise exposure that induces auditory neuropathy caused by deterioration of HCs, loss of ribbon synapses between IHCs and ANFs, or damaged ANFs without functional elevation of hearing thresholds based on ABR analysis (Paquette et al., 2016). However, using proteomics, we found that even loud noise exposure can have broad effects on cochlear structures and cells (Figure 3). We also found changes in protein abundance, indicating disruption of membranes and cell junctions, damage to stereocilia, and altered synapses (Figure 1). We anticipate that damage, excitotoxicity, and the associated stress likely impair numerous protein-protein interactions, drive protein misfolding, and contribute to protein accumulation in numerous cell types. However, our bulk cochlear proteomic and transcriptomic analyses are likely blinded to some responses because of molecular averaging among cell types and limited sensitivity.
We used two complementary, relative, and quantitative proteomic methods to investigate how acoustic overstimulation drives hearing loss. We also used LC-MS/MS to verify an absolute increase in the level of Tpm1 and reduction of Col1a1. Metabolic stable isotope labeling with 15N or heavy amino acids provides an accurate method with a broad dynamic range for measuring changes in relative protein abundance both in vitro (Zhang et al., 2009) and in vivo (Gouw et al., 2010; Savas et al., 2015, 2017), but it is limited by measurement of only partially overlapping pools of peptides in each MS analysis run. Isobaric peptide labeling strategies (i.e., TMT) are also powerful, because sixteen or more samples can be multiplexed and analyzed in the same MS analysis run (Frost and Li, 2016; Weekes et al., 2014). However, TMT analysis suffers from ratio compression. We confirmed many trends in protein FCs obtained from the 15N analyses with TMT-based measurements. The two strategies generally agreed but were admittedly unsuccessful in precisely reproducing the same quantitative measures. Similar to other high-throughput analysis methods, MS has intrinsic technical challenges and biases (Bakalarski and Kirkpatrick, 2016; Schubert et al., 2017). Confounding factors include biological sources such as animal-to-animal variation, uncontrollable experimental differences during sample processing, technical limitations, and differences in the bioinformatic analysis (Rauniyar and Yates, 2014). Altogether, these factors limit, but do not prevent, our ability to directly compare and confirm our proteomic measures.
Quantifying cochlear protein abundance presents numerous technical challenges. First, we are sample limited in all of our cochlear studies, because a single animal provides only tens of micrograms of protein. Thus, some of our experiments required pooling of extracts from multiple animals, which increases the complexity of our samples and ultimately reduces the sensitivity of our analyses. Second, we found that there is an inherent variability in how different animals respond to relatively short noise exposure. Overall, we found that anesthetized mice exposed to 100 dB SPL for 30 min represent a reliable model to study the effect of loud noise exposure with the caveat that thresholds do not fully recover. Awake exposure to 94 dB SPL for 30 min produces fully recoverable thresholds, but this model is limited by some variability because of noise avoidance behaviors (Figure S1). Altogether, our models provide complementary lenses to view and study the temporary aspects of NIHL.
Proteotoxicity represents a disequilibrium in protein homeostasis because of translational errors, protein misfolding, and oxidative stress, which overwhelms the chaperone and protein degradation machinery (Balch et al., 2008; Morimoto, 2008). Chronic disequilibrium is likely to drive the proteostatic collapse often seen in neurodegeneration, cancer, metabolic diseases, and aging (Morimoto, 2011). Heat shock proteins (HSPs) in particular have been previously found to be selectively elevated in the cochlea after noise exposure, presumably playing protective roles (Gong et al., 2012; Lim et al., 1993; Yoshida et al., 1999). Our results support and extend these findings, because elevated levels of Hspa1a (Hsp72), Hspa1b (Hsp70), and the cytosolic HSPs Hsp90aa1 and Hsp90ab1, as well as Hsp90b1 localized to the endoplasmic reticulum, nucleus, plasma membrane, and vacuole, were found to be altered after noise exposure (Figure 1; Table S1). The heat shock response has been shown to be non-uniform in its severity from cell to cell following stress, which is thought to be protective at the population level (Santiago et al., 2020). We observed instances of non-uniform HSP90b1 levels across IHCs, potentially highlighting the complexity of noise-induced stress at the cellular level (Figure 4). A protective role for elevated HSP levels in the human cochlea is supported by the observation that the perilymph from cochlear implant patients with residual hearing loss compared with complete hearing loss had selectively elevated HSP abundance (Schmitt et al., 2018), suggesting that our findings of elevated HSP levels in our mouse models of NIHL may potentially underlie critical mechanisms in the preservation of human hearing. Hsp90aa1, which we have shown to have increased FCs following noise exposure, was also elevated in human patients with impaired hearing. The presence of Hsp90aa1, which is secreted into the perilymph during oxidative stress, and details of its protective role in hearing loss remain unclear.
As far as we are aware, this is the first discovery of elevated levels of the proteasome after noise exposure. Previous studies have focused almost exclusively on characterizing the cochlear transcriptome after noise exposure (Maeda et al., 2017; Yang et al., 2015, 2016). Importantly, we recapitulated many of these findings in our characterization of the noise-exposed cochlear transcriptome. Despite exposing mice to shorter periods of lower-intensity noise, i.e., 105 dB SPL for 30 min versus 120 dB SPL for 1 h, we observed the same trend (i.e., more genes with elevated rather than decreased levels) (Yang et al., 2015, 2016). Protein posttranslational modifications within the cochlea have largely been studied under the lens of cellular development, leaving the importance of glycosylation, ubiquitylation, and SUMOylation within the context of NIHL poorly characterized (Mateo Sánchez et al., 2016). Although preliminary, our findings provide the first proteome-wide analysis of cochlear protein ubiquitylation following noise exposure. The importance of ubiquitylation in NIHL is supported by findings from ubiquitin-specific protease 53 (Usp53) mutant mice (Kazmierczak et al., 2015). Usp53, a member of the deubiquitinating enzyme family, localizes to tight adherens junctions surrounding OHCs and supporting cells. Loss-of-function Usp53 mutant mice have increased susceptibility to noise injury at high frequencies, which is believed to result from a weakening of the tight adherens junctions (Kazmierczak et al., 2015). We discovered several collagen proteins that provide structural support within the cochlea, with reduced FCs after exposure to loud or very loud noise. We hypothesize that these structures are damaged by excess noise, become ubiquitylated, and are subsequently degraded. This is supported by our diGly MS results, which identified Col16a1 as a protein ubiquitylated after exposure to very loud noise. Altogether, this potentially highlights the importance of ubiquitylation with respect to NIHL and the need for a more robust investigation into the role of these ubiquitylated proteins following noise exposure.
In summary, we found that auditory overstimulation acutely unbalances the cochlear proteome by driving the accumulation of hundreds of proteins. This disequilibrium activates the proteostasis network by elevating protein chaperones and proteasome levels in multiple inner ear cell types. Most proteins identified in the investigation have never been previously linked to acoustic injury or associated stress response. Therefore, this research provides a path for future studies aimed at verifying their functional involvement in the cochlear stress response to acoustic overstimulation and the process of hearing threshold recovery.
STAR★METHODS
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Jeffrey N Savas (jeffrey.savas@northwestern.edu).
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
The accession number for the MS data reported in this paper is MASSIVE: MSV000084739 and ProteomeXchange: PXD016905. The accession number for the RNA-seq data reported in this paper is GEO: GSE160639.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
This study used FVB (001800 - FVB/NJ - The Jackson Laboratory) mice, 7–8 weeks old, with equal number of males and females. All animal experiments were conducted according to Veterans Administration San Diego Healthcare System, University of California–San Diego Institutional Animal Care and Use Committee (IACUC) TSRI IACUC / Department of Animal Resources (approved protocol number 07–0083), Northwestern University IACUC (approved protocol numbers IS00001182 and IS00011904).
METHOD DETAILS
Noise exposures and cochlear functional assays
Noise exposures were delivered to anesthetized (Ketamine (100 mg/kg) and xylazine (3 mg/kg)) or unanesthetized animals held within small wire cages in a custom-built sound-proof chamber designed by Charles Liberman (Mass. Eye and Ear). The box is constructed of 3/4” plywood sheets. The basic principle is that no two sides are parallel. The front and back panel are the same, except that the back is truncated at a height of 42′’ (rather than 48”) and has no door. This makes the top slanted with respect to the floor. The side panels are then cut to fit. The top panel has a rectangular hole cut in it to which an exponential horn and the acoustic driver are mounted.
Waveforms used for acoustic overexposure were designed using a waveform generator (Tucker-Davis Technologies, Alachua, FL) within the frequency range of 8–16 kHz for 30 min. Noise level intensity was delivered through an exponential horn extending from compression driver (JBL, 2446H/J, Northridge, CA). The mice randomly assigned to the noise exposure groups, and exposed to ambient (65 – 70, or 70 dB SPL), loud (94 or 100 dB SPL), or very loud (105 dB SPL). Noise generation stability was monitor by PicoLog system (Picoscope 2000 series) and calibrating microphone (PCP Piezotronics, NY).
Auditory brainstem responses (ABRs) and distortion product otoacoustic emission (DPOAE) were recorded in young adult FVB mice (P50–60). Baseline hearing measurements were recorded from 7–8 week old animals and sequentially recorded 1, 7 and 14 days after noise exposure. ABR and DPOAE recordings were obtained by Tucker Davis Technologies (TDT) System III workstation running BioSigRP in a soundproof chamber (EcKel audiometric room, Cambridge, MA). ABR stimulus were pre-amplified with a 4-channel MEDUSA pre-amp and analyzed by a real-time processor in TDT system3. Prior to electrode placement, animals were anesthetized by intraperitoneal injection of Ketamine and Xylazine cocktails given at a dose of 100 mg/kg and 3 mg/kg body weight, respectively. The top-up injections (quarter of the original dose) were administered as needed. Animal body temperature was maintained at 37–38°C by electric heating pad (Homeothermic blanket system, Harvard Apparatus). The ABR thresholds upon click and tone burst stimuli were recorded with subcutaneous platinum needle electrodes placed at the vertex (recording) and ipsilateral mastoid (reference), with the ground electrode placed on the lower back. ABR click stimulus was set at 0.1 ms of click duration with the rate of 20 Hz. The stimulus intensities were ranged from 15 dB SPL to 80 dB SPL with 5 dB increment steps. Tone ABRs was established by 20 ms tone pips, ranging from 8–32 kHz with 4 kHz increments and presented from 20 to 80 dB SPL stimulus level with 10 dB step. An average of 500 stimuluses presentation was displayed on a PC monitor during the experiments using operating software (BioSigRP, TDT). ABR thresholds were defined as the lowest stimulus for which recognizable ABR waves could be observed. The amplitude of ABR wave-I was estimated by measuring the voltage difference between the wave-I peak and the trough between wave-I and wave-II. DPOAE recording was verified by using Etymotic low noise microphone system ER-10B+. The stimulus which consist of two primary pure-tone frequencies (f1 and f2) was synthesized differing by a factor of 1.2. The L1 and L2 values were varied from 15 to 75 dB SPL (L1 > L2, 5 dB) in 5 dB step across the frequency from 8–32 Hz. The spectral magnitude of the two primaries, 2f1 – f2 distortion product, and the noise floor will be determined from an averaging of 500 stimuli.
Metabolic labeling of the cochlea with 15N mouse chow
12 FVB mice were metabolically labeled with 15N-rich, Spirulina-based diet (Cambridge Isotopes and Harlan Laboratories) for two generations as we have previously described (Butko et al., 2013). The second generation “heavy” mice were maintained on the 15N-rich, Spirulina-based diet until P40–50 at which time the cochlea were harvested and stored at −80°C until homogenization and pooling. The 15N protein enrichment was determined to be greater than 95% in the brain (MacCoss et al., 2003).
Preparation of cochlear extracts for 15N based quantitation
FVB mice were randomly assigned into 5 groups (4 mice per group), 15N heavy labeled control, and 4 experimental groups of normal 14N diet (0 dB, 70 dB, 100 dB and 105 dB). Mice were sacrificed and the cochlea was immediately dissected from temporal bones and the bony/cartilaginous capsule and vestibular sensory organs (organ of Corti, utricular and saccular maculae, and semicircular canal ampullae) were removed by microdissection in less than ten minutes after the end of noise exposure and euthanasia. The extracts were homogenized in 4 mM HEPES, 0.32 M sucrose, that was amended with protease inhibitors. The 14N cochlear homogenate was then mixed 1:1 with 15N homogenate and centrifuged at 1,000 × g for 10 min (4°C). The pellet was discarded, and supernatant was subsequently centrifuged at 10,000 × g for 15 min (4°C) to collect the crude protein pellet used for mass spectrometry. Protein concentration was verified, and 8 M urea was added. 100 μg of extracted protein was taken from each sample and subsequently processed with ProteasMAX (Promega) according to manufacturer’s instructions. Disulfides within the samples were selectively reduced by 5 mM Tris (2 carboxyethyl) phosphine at room temperature for 20 min, alkylated in the dark by 10 mM iodoacetamide (IAA) for 20 min, and trypsinized (Sequencing Grade Modified Trypsin, Promega) overnight at 37°C, trypsinization was quenched by acidification.
MudPIT and LTQ Velos Orbitrap MS analysis
Digested proteins were pressure-loaded into a 250 μm i.d capillary packed with 2.5 cm of 10 μm Jupiter C18 resin (Phenomenex) followed by an additional 2.5 cm of 5-μm Partisphere strong cation exchanger (Whatman). The buffer containing 95% (vol/vol) water, 5% (vol/vol) acetonitrile, and 0.1% formic acid was used to wash the column. An i.d capillary (100 μm) with Jupiter C18 resin (15 cm,4 μm) attached with a pulled tip packed (5 μm) and the entire split column (desalting column–union–analytical column) was placed in line with an Agilent 1200 quaternary HPLC and analyzed using a modified 11-step separation (Link et al., 1999; Washburn et al., 2001). Three buffer solutions were used in the following step, buffer A (5% (vol/vol) acetonitrile/0.1% formic acid), buffer B (80% (vol/vol) acetonitrile/0.1% formic acid) and buffer C (500 mM ammonium acetate/5% (vol/vol) acetonitrile/0.1% formic acid). Step 1 consisted of a 90-min gradient from 0%–100% (vol/vol) of buffer B. Steps 2–11 had a similar profile with the following changes: 5 min in 100% (vol/vol) of buffer A, 3 min of buffer C in 10, 20, 30, 40, 50, 60, 70, 80, 90, and 100% (vol/vol), respectively, for the 11-step analysis, a 10-min gradient from 0 to 15% (vol/vol) buffer B, and finally a 108-min gradient from 15%–100% (vol/vol) buffer B. Eluted peptides from microcapillary column were electrosprayed into an LTQ Velos Orbitrap mass spectrometer (Thermo Finnigan) with the application of a distal 2.4-kV spray voltage. A cycle of one full-scan mass spectrum was set at a resolution of 60,000 (400–1,800 m/z) followed by 15 data-dependent MS/MS spectra at a 35% normalized collision energy was repeated continuously throughout each step of the multidimensional separation. MS1 maximum ion accumulation times were set to 500 ms and 100 ms for MS2 scans. Charge state rejection was set to omit single charged ion species and ions for which a charge state could not be determined for MS/MS. Signal for fragmentation was set to 1,000 as a minimal level. Dynamic exclusion was enabled with just single repeat count at a duration of 20.00 s, list size of 300, exclusion duration of 30.00 s, and exclusion mass with high/low of 1.5 m/z. Mass spectrometer scan functions and HPLC solvent gradients were controlled by the Xcaliber data system.
Preparation of cochlear extracts for TMT based quantitation
FVB mice were randomly assigned to 94 dB or 105 dB noise exposure groups. Cochleae were collected either immediately or 2 weeks after noise exposure. Animals were euthanized and the cochlea was micro dissected as described prior to further homogenization via Precellys 24 (Bertin Technology, Rockville, MD) in Syn-PER (Thermo Scientific) lysis buffer amended with protease inhibitor cocktail. Protein was separated from impurities and lipids by methanol-chloroform precipitation prior to resuspension in 6M guanidine in (50 mM HEPES). Proteins were further processed via the reduction of disulfide bonds with DTT, and alkylation of cysteine residues with iodoacetamide. Proteins were then digested for 3 h with 2 mg of endoproteinase LysC (Promega) and subsequently digested overnight with 2 μg of Trypsin (Pierce). The digest was then acidified with formic acid to a pH of ~2–4 and desalted by using C18 HyperSep cartridges. The eluted peptide solution was completely dried before verifying the concentration by micro BCA assay (Thermo Scientific, Rockford, IL). Equal concentration of peptide (~100 μg) from each sample was then used for isobaric labeling. TMT-10 or 15plex labeling on peptide samples were performed according to the manufacturer’s instructions (ThermoFisher Scientific). After 2 h incubation at room temperature, the reaction was quenched with hydroxylamine at a final concentration of 0.3% (v/v). Isobaric labeled samples were then combined 1:1:1:1:1:1:1:1:1:1 or combined 1:1:1:1:1:1:1:1:1:1:1:1:1:1:1 and lyophilized. The combined isobaric labeled peptide samples were then fractionation by Hypersep SCX SPE column and eluted by the following gradient concentrations of NH4AcO: 20 mM, 50 mM, 100 mM, 500 mM, 1,000 mM and 2,000 mM. Fractionated samples were then desalted by loose resin tips (ImcsTipS, RP-20mg) and the concentration was again determined by Micro BCA. Peptide solutions were dried, stored at −80°C, and reconstituted in LC-MS buffer A (5% acetonitrile, 0.125% formic acid) until LC-MS/MS analysis.
Nanoflow LC with multinotch MS2/MS3 Orbitrap Fusion MS analysis
Three micrograms from each fraction were loaded for LC-MS analysis using a Thermo Orbitrap Fusion coupled to a Thermo EASY nLC-1200 UPLC pump and vented Acclaim Pepmap 100, 75 μm × 2 cm nanoViper trap column and nanoViper analytical column (Thermo–164570), 3 μm, 100 Å, C18, 0.075 mm, 500 mm with stainless steel emitter tip assembled on the Nanospray Flex Ion Source with a spray voltage of 2000V. The chromatographic run was performed by 4 h gradient beginning with 100% buffer A (5% acetonitrile, 0.125% formic acid), 0% B (95% acetonitrile, 0.125% formic acid) and increased to 7% B over 5 mins, then to 25% B over 160 mins, 36% B over 40 mins, 45% B over 10 mins, 95% B over 10 mins, and held at 95% B for 15 mins before terminating the scan. Multinotch MS3 method (McAlister et al., 2014) was programmed as the following parameter: Ion transfer tube temp = 300 °C, Easy-IC internal mass calibration, default charge state = 2 and cycle time = 3 s. MS1 detector set to orbitrap with 60 K resolution, wide quad isolation, mass range = normal, scan range = 300–1800 m/z, max injection time = 50 ms, AGC target = 2 × 105, microscans = 1, RF lens = 60%, without source fragmentation, and datatype = positive and centroid. Monoisotopic precursor selection was set to included charge states 2–7 and reject unassigned. Dynamic exclusion was allowed n = 1 exclusion for 60 s with 10ppm tolerance for high and low. An intensity threshold was set to 5 × 103. Precursor selection decision = most intense, top speed, 3 s. MS2 settings include isolation window = 0.7, scan range = auto normal, collision energy = 35% CID, scan rate = turbo, max injection time = 50 ms, AGC target = 1 × 104, Q = 0.25. In MS3, the top ten precursor peptides were selected for analysis were then fragmented using 65% HCD before orbitrap detection. A precursor selection range of 400–1200 m/z was chosen with mass range tolerance. An exclusion mass width was set to 18 ppm on the low and 5 ppm on the high. Isobaric tag loss exclusion was set to TMT reagent. Additional MS3 settings include an isolation window = 2, orbitrap resolution = 60 K, scan range = 120 – 500 m/z, AGC target = 1*104, max injection time = 120 ms, microscans = 1, and datatype = profile.
Absolute quantitation of peptide / protein abundance
Synthetic peptides were synthesized containing a single heavy C-terminal arginine residue (JPT Peptide Technologies GmbH, Berlin, Germany). SpikeTide TQL peptides were quantified using a proprietary Quanti-Tag. Peptides are released from tag by trypic digestion and aliquoted at nM. These peptides were spiked (0.5 nM) into cochlear peptide mixtures from three independent biological replicates (prepared as described above) each exposed to 70 dB SPL, 94 dB SPL, and 105 dB SPL and purified with C18 Ziptips. The purified peptide concentrated using a SpeedVac vacuum concentrator (Labconco Inc.), and analyzed by shotgun LC-MS/MS with a 2 or 4 hour analysis runs with an Orbitrap Fusion MS as described above. The resulting spectra was extracted, searched, and quantified as described below with Prolucid/Sequest DTASelect and Census. We used the reconstructed MS1 chromatograms (relative area under the curves) to determine the absolute peptide quantities.
Analysis of tandem mass spectra
The Integrated Proteomics Pipeline - IP2 (Integrated Proteomics Applications, Inc., http://www.integratedproteomics.com/) was used to analyze the proteomic results with ProLuCID, DTASelect2, Census, and QuantCompare. Spectrum raw files were extracted into ms1 and ms2 files by using RawExtract 1.9.9 software (http://fields.scripps.edu/downloads.php) (McDonald et al., 2004). Tandem mass spectra were searched against the European Bioinformatic Institute (IPI) mouse protein database (https://www.ebi.ac.uk/IPI/IPImouse.html). The target/decoy database containing the reversed sequences of all proteins was used to accurately determine peptide probabilities and FDRs (Peng et al., 2003a). ProLuCID searches on an Intel Xeon cluster was used to match tandem mass spectra to peptide sequences with 50 ppm peptide mass tolerance for precursor ions and 400 ppm for fragment ions (Xu et al., 2015). The search space included fully and half-tryptic peptide candidates that matched within the mass tolerance criteria with no miscleavage constraint. Carbamidomethylation (+57.02146 Da) of cysteine was considered as a static modification. DTASelect was used to access validity of peptide/spectrum matches (PSMs) (Cociorva et al., 2006; Tabb et al., 2002) by using two SEQUEST-defined parameters (Eng et al., 1994), the cross-correlation score (XCorr), and normalized difference in cross-correlation scores (DeltaCN). The search results were grouped by charge state (+1, +2, +3, and greater than +3) and tryptic status (fully tryptic, half-tryptic, and non-tryptic), resulting in 12 distinct subgroups. The distribution of Xcorr, DeltaCN, and DeltaMass values for (a) direct and (b) decoy database PSMs in each of these subgroups, was obtained. The direct and decoy subsets were then separated by discriminant analysis. Peptide match probabilities were calculated based on a nonparametric fit of the direct and decoy score distributions. The minimum threshold peptide confidence was set at 0.95. The FDR was then determined as the percentage of reverse decoy PSMs among all the PSMs which passed the confidence threshold. The minimum of one peptide was required for each protein identification, this peptide had to be an excellent match with an FDR less than 0.001 and at least one excellent peptide match. The quantified proteins which FDRs below 1% were estimated for each sample analysis. For the 14N / 15N analysis, each of protein dataset was searched twice in the light and heavy protein SEQUEST databases, respectively. The search results were then filtered using DTASelect2 and ion chromatograms were generated using an updated version of a program previously written in our laboratory (MacCoss et al., 2003). This software, called “Census,” is available from the authors for individual use and evaluation through an Institutional Software Transfer Agreement (see http://fields.scripps.edu/yates/wp/?page_id=824 and http://fields.scripps.edu/download/Census.tar.gz for details) (Park et al., 2008). First, the elemental compositions and corresponding isotopic distributions for both the unlabeled and labeled peptides were calculated. This information was then used to set an appropriate m/z range from extracted ion intensities, which included all isotopes greater than 5% of the calculated isotope cluster base peak abundance. MS1 files were used to generate chromatograms from the m/z range surrounding both the unlabeled and labeled precursor peptides. Census calculates peptide ion intensity ratios for each pair of extracted ion chromatograms. The linear least-squares correlation was used to calculate the ratio (i.e., slope of the line) and closeness of fit [i.e., correlation coefficient (r)] between the data points of the unlabeled and labeled ion chromatograms. Finally, QuantCompare was used to analyze protein ratios which represent by log two-FC and ANOVA p value with BH correction to identify significant quantified proteins.
For TMT, the raw spectral raw files were extracted into MS1, MS2, and MS3 files using the in-house program RawConverter (He et al., 2015). Spectral files were then pooled from fractions for each sample and searched against the Uniprot mouse protein database and matched to sequences using the ProLuCID/SEQUEST algorithm (ProLuCID ver. 3.1) with 50 ppm peptide mass tolerance for precursor ions and 600 ppm for fragment ions. Fully and half-tryptic peptide candidates were included in search space, all that fell within the mass tolerance window with no miscleavage constraint, assembled and filtered with DTASelect2 (ver. 2.1.3) through the Integrated Proteomics Pipeline (IP2 v.5.0.1, Integrated Proteomics Applications, Inc., CA, USA). Static modifications at 57.02146 C and 229.162932 K and N-term were included. The target-decoy strategy was used to verify peptide probabilities and false discovery ratios (Elias and Gygi, 2007). Minimum peptide length of five was set for the process of each protein identification. Each dataset had an 1% FDR rate at the protein level based on the target-decoy strategy.Isobaric labeling analysis was established with Census 2 as previously described (Park et al., 2014). TMT channels were normalized by dividing it over the sum of all channels. No intensity threshold was applied. The fold change was then calculated as the mean of the experimental group standardized values. p-values were then calculated by Student’s t test with the B.H. adjustment.
Bioinformatic analysis with PATHER AND STRING
Protein ontologies were investigated with the protein analysis through evolutionary relationships (PANTHER) system (http://www.pantherdb.org), in complete cellular component and complete molecular function categories (Mi et al., 2013, 2016). The statistical overrepresentation test was calculated by using the significant proteins from each individual noise exposure experiment as the query and the aggregated total proteins identified in all three noise exposure conditions as the reference. Protein ontologies with Fisher statistical tests with false discovery rate (FRD) correction less than 0.05 were considered significant.
The Search Tool for the Retrieval of Interacting Genes (STRING) database was used to determine protein-protein interaction from significant quantified protein which found in each noise exposure condition. The STRING resource is available at http://string-db.org (Szklarczyk et al., 2017). The corresponding protein–protein interaction networks were constructed with a highest confidence of interaction score at 0.9.
Heatmaps, clustering and volcano plots for the recovery period
Reporter ion intensity from the MS/MS analysis were analyzed using NCI-BRB ArrayTool software. The data was normalized using quantile normalization and differentially expressed (DE) proteins were selected based on univariate t test with a p value cut off 0.05 and minimum fold change on 1.5. The selected DE proteins were used to create heatmaps using the “Dynamic Heatmap Viewer” module implemented in the ArrayTool using default parameters, distance metrics of one-minus correlation and average linkage. Volcano plots were created in the R statistical program using ggplot2 with log2 fold change on the x axis and −log10 p value on the y axis.
RNaseq and transcriptomic quantification
Total RNA from cochlea were extracted using Trizol as per the manufacturer’s recommended protocol, and further purified via RNeasy Plus Mini kit (QIAGEN). Purified RNA was sent to Novogene (Novogene Corporation Inc, Sacramento, CA) for QC, library preparation, sequencing, and quantitation as per the in-house standard protocol for gene expression analysis. RNA degradation and contamination was monitored on 1% agarose gels. RNA purity was determined by NanoPhotometer® spectrophotometer (IMPLEN, CA, USA). RNA integrity and quantitation were assessed using the RNA Nano 6000 Assay Kit of the Bioanalyzer 2100 system (Agilent Technologies, CA, USA). The samples passing the QC step were then used for library preparation and sequencing.
One microgram of RNA per sample was used as starting material for sequencing library generation. NEBNext® UltraTM RNA.Library Prep Kit for Illumina® (NEB, USA) was used for library generation following manufacturer’s recommendations and index codes were added to attribute sequences to each sample. Briefly, mRNA was purified from total RNA using poly-T oligo-attached magnetic beads. Fragmentation was carried out using divalent cations under elevated temperature in NEBNext First Strand Synthesis Reaction Buffer (5X). First strand cDNA was synthesized using random hexamer primer and M-MuLV Reverse Transcriptase (RNase H-). Second strand cDNA synthesis was subsequently performed using DNA Polymerase I and RNase H. Remaining overhangs were converted into blunt ends via exonuclease/polymerase activities. After adenylation of 3′ ends of DNA fragments, NEBNext Adaptor with hairpin loop structure were ligated to prepare for hybridization. To select cDNA fragments of preferentially 150~200 bp in length, the library fragments were purified with AMPure XP system (Beckman Coulter, Beverly, USA). Then 3 μL USER Enzyme (NEB, USA) was used with size-selected, adaptor ligated cDNA at 37°C for 15 min followed by 5 min at 95°C before PCR. Then PCR was performed with Phusion High-Fidelity DNA polymerase, Universal PCR primers and Index (X) Primer. At last, PCR products were purified (AMPure XP system) and library quality was assessed on the Agilent Bioanalyzer 2100 system. The clustering of the index-coded samples was performed on a cBot Cluster Generation System using PE Cluster Kit cBot-HS (Illumina) according to the manufacturer’s instructions. After cluster generation, the library preparations were sequenced on an Illumina Novaseq6000 and paired-end (150 bp) reads were generated.
Raw data (raw reads) of FASTQ format were processed through fastp. In this step, clean data (clean reads) was obtained by removing reads containing either adaptor or poly-N sequences and/or reads with low quality scores. At the same time, Q20, Q30 and GC content of the clean data were calculated. Reference genome and gene model annotation files were downloaded from genome website browser (NCBI/UCSC/Ensembl) directly. Indexes of the reference genome was built using STAR and paired-end clean reads were aligned to the reference genome using STAR (v2.5). STAR utilizes the Maximal Mappable Prefix (MMP) method which can generate a precise mapping result for junction reads. HTSeq (v0.6.1) was used to count the read numbers mapped to each gene. We obtained greater than 75 million mapped reads at a total mapping rate > 95.3% from each of the four biological replicates per group. We obtained differential gene expression profiles for 13,519 and 16,258 transcripts for loud versus ambient and very loud versus ambient datasets respectively.
Read count per transcript were used for further analysis with the iGEAK program, an interactive gene expression analysis kit using the R/shiny platform. Read counts were filtered to remove very low signals (i.e., we required a minimum read count = 8 in at least 4 samples). Data were normalized using edgeR’s TMM (Trimmed Mean of M-values) normalization method. EdgeR method provides statistical routines for determining differential expression in digital gene expression data, and the resulting P values were adjusted using the Benjamini and Hochberg’s approach for controlling the False Discovery Rate (FDR). Genes with an adjusted P value < 0.05 and fold change > 1.5 found by edgeR were assigned as differentially expressed genes. Principal Component Analysis (PCA) plots, Correlation (Pearson correlation) plots, and Volcano plots were also created using iGEAK program.
TUBE pulldown western blot
Mice were randomly assigned to either the ambient, loud, or very loud noise exposure group (5 mice per group) and exposed for 30 minutes prior to the harvesting of cochlea. Cochlea from each group were then pooled respectively (total of 10 cochlea per noise exposure group) prior to homogenization via Precellys 24 in modified lysis buffer: 50 mM Tris-HCl (pH 7.5), 0.15 M NaCl, 1 mM EDTA, 1% NP-40, 10% glycerol, Ub/Ub protease inhibitors, PR-619 and 1,10-phenanthroline (o-PA). Protein concentrations were measured via micro BCA and 1.0 mg of cochlear homogenate were used for the isolation of polyubiquitylated proteins via Magnetic beads coated with Tandem Ubiquitin Binding Entities (TUBEs). Briefly, 100 μL of beads were washed with TBST and then incubated with cochlear homogenates at 4°C for 4 hours. After binding, protein homogenate was removed from the beads, and the beads were washed three times with TBST. Bound ubiquitinated proteins were eluted by boiling the beads in 100 μL of SDS prior to incubation of supernatant in reducing sample buffer for 5 minutes. This solution was then centrifuged at 13,000xg for 5 minutes at 4°C, 40 μL of each sample was then loaded into a 5%–15% Tris gel in addition to input and positive control (10.0 μL of MG132 treated HEK293T cell lysate). Following gel electrophoresis, the proteins were transferred to nitrocellulose via wet transfer method in transfer buffer with 0.02% SDS. The blots were blocked with 15 mL of 1X sigma block for 2 hours, and then incubated with primary antibody, 1:1000 anti-Ubiquitin Antibody (P4D1, sc-8017), overnight at 4°C. Blots were washed three times with TBST, and then incubated with anti-mouse IgGHRP (1:5000) for 1 hour at room temperature. Blots were washed three times with TBST, and the immune reaction was then assed using ECL, and imaged via Chemi doc gel imaging system (Bio-rad).
diGly anlaysis of protein ubiquitination
Mice were exposed to Noise and cochlea were harvested as described above (5 mice per condition). Cochlea were pooled (10 cochlea per condition) and soluble protein was extracted using Urea cell lysis buffer containing 20 mM HEPES (pH 8.0), 9 M Urea, 1mM Sodium orthovandate, 2.5 mM sodium pyrophosphate,1 mM b-glycerophosphate and homogenized by Percellys. Cochlear lysates were then further homogenized via needle sonication, lysates were then incubated on ice for 10 minutes prior to centrifugation at 10,000 g for 10 minutes. Supernatant was then collected and digested for diGly peptide capture with PTMScan Ubiquitin Remnant Motif Kit (cat# 5562, Cell Signaling technology, USA) as per recommended protocol. Briefly, cochlear lysates were reduced with 5 mM DTT and alkylated with idoacetaminde solution. Urea concentration was reduced via the addition of 20 mM HEPES to a final concentration of 2 M Urea prior to protein digestion with LysC overnight at 37°C. The following day, trypsin was added at a concentration of 2 μg/100 μg and incubated overnight at 37°C. Peptides were then desalted and purified using Hypersep columns and dried down. 400 μg of purified peptides were then solubilized in 1.4 mL of Immunoaffinity Purification buffer (IAP). DiGly bead slurry was washed four times with PBS and added to the solubilized peptide solutions for overnight incubation at 4°C with end over end rotation. Beads were separated from supernatant by centrifugation and washed one time with IAP buffer and then three times with chilled DI water. Bound peptides were eluted from the beads by incubating with 40 μL of 0.15% TFA for 10 min at room temperature, this was repeated once and both elutions were pooled. Eluted diGly peptides were desalted using spin columns and dried. Dried peptides were resolubilized in MS loading buffer contain 5% acetonitrile and 0.1% TFA and injected for MS/MS analysis.
Immunofluorescence (IF) with confocal microscopy analysis
Immediately following noise exposure, FVB mice were anesthetized and cochlea were collected. Oval and round windows were punctured and a small hole was created near the apex of the cochlea for perfusion with 4% paraformaldehyde for 3 hours at 4°C. For the mid-modiolar sections, Cochlea were decalcified in Immunocal (Decal Chemical Corporation, Congers, NY) for 8 hours prior to cryoprotection for 2 days in 30% sucrose. Cochlea were mounted in OCT and sectioned at 12 μm for IF. Slides were rinsed three times in PBS at room temperature followed by blocking in 20% NHS 0.5% Triton-X (Cat# 1086431000, Millipore) for 1 hour at room temperature prior to primary antibody incubation (see below). For cochlea whole mount, cochlea were decalcified in Immunocal (Decal Chemical Corporation, Congers, NY) overnight at 4°C or until cochlea became translucent. The decalcified bone, lateral wall, Reissner’s membrane and tectorial membranes were dissected away from the organ of Corti. Whole mounted cochlea were then sectioned into 4 pieces and cryoprotected in 30% sucrose for 30 minutes prior to freezing on dry ice. The frozen pieces were then thawed and rinsed three times in PBS for 10 minutes at room temperature with agitation. Cochlear sections were then blocked for 1 hour in 20% NHS 1% Triton-X blocking buffer at room temperature. Primary antibodies were diluted in blocking buffer as follows: Recombinant Anti-ARPC2 antibody (1:100, Abcam, ab133315), Human gp96/HSP90B1 90b1 (1:200, R&D Systems, MAB7606, 816803), Anti-LTBP4 antibody (1:100, Abcam, ab222844), COL9A1 Polyclonal Antibody (1:100, Invitrogen, PA5–93062), Neurofilament H (1:1000, Synaptic Systems, 171 106,), Myosin-V11A (1:500, Proteus Bioscience, 25–6790, RRID:AB_1001525), PSMC5 (1:200, Cell Signaling, 13392S), VCP (1:200, Abcam, ab111740), UBA2 (1:200, Cell Signaling, 5293) and incubated overnight at 4°C for sections and at 37°C for whole mounts. Following primary antibody incubation slides were rinsed three times with PBS for 15 minutes at room temperature. Slides were incubated in secondary antibody: 1:500 Goat Anti-Mouse IgG H&L (Alexa Fluor® 488, Abcam, ab150113), 1:500 Goat Anti-Rabbit IgG H&L (Alexa Fluor® 568, Abcam, ab150088), 1:50 Alexa Fluor 647 Phalloidin (Thermo Fisher, A22287) for three hours at room temperate. Slides were then rinsed three times in PBS for 15 minutes and stained with DAPI Staining Solution (Abcam, ab228549) for 5 minutes prior to mounting in Fluoromount-G (Thermo Scientific, cat. 00–4958-02). Tissues were counterstained with DAPI nucleic acid stain and/or Alexa Fluor-conjugated phalloidin actin stain. Images were captured with confocal laser scanning microscopy (Leica DMI4000) with identical settings. Images were cropped and the brightness contrast was adjusted with ImageJ software using constant settings.
Synapse density measurements
For synapse quantification, immediately following noise exposure to 70 dB, 94 dB or 105 dB, FVB mice were euthanized and perfused with ice cold 4% paraformaldehyde (pH 7.4) until exsanguinated. The cochlea were then micro dissected as described above and incubated at 4°C in 4% paraformaldehyde (pH 7.4) for 3 hours prior to decalcification with Immunocal overnight at 4°C. Cochlea were micro dissected as described in the above section prior to permeabilization with 2% Titon-X-100 in PBS for 30 minutes at room temperature. Samples were then rinsed with PBS for 5 minutes prior to blocking for two hours in 20% NHS. Cochlea were incubated overnight at 37°C in primary antibody solution containing CtBP2-ms-IgG1 (1:200, BD Transduction, 612044), and GluR2-ms-IgG2a (1:2000, Millipore, MAB397). Samples were rinsed 3 times for 10 minutes with PBS then incubated for 2 hours at 37°C with secondary antibodies: 1:1000 Goat Anti-Mouse IgG2a Cross-Adsorbed Secondary Antibody (Alexa Fluor® 488, Invitrogen, A-21131), and 1:1000 Goat Anti-Mouse IgG1 Cross-Adsorbed Secondary Antibody (Alexa Fluor® 568, Invitrogen, A-21124). Following three 15-minute washes with PBS, cochlea were stained with DAPI and mounted with Fluoromount-G. Images were captured with confocal laser scanning microscopy (Leica DMI4000) with identical settings. Cochlear pieces were measured from base to apex and regions corresponding to the 9–12, 12–16 and 24–28 kHz regions of the cochlea were captured for synapse density measurements. Images were cropped and the brightness contrast was adjusted with ImageJ software using constant settings. Areas in which presynaptic CtBP2 puncta overlapped with postsynaptic GluR2 puncta were then counted as a single synapse.
SEM analysis
FVB mice were euthanized and perfused with ice cold 4% paraformaldehyde in 0.1 M sodium cacodylate, pH 7.4 until exsanguinated immediately following noise exposure. All buffers were prepared and filtered using PES 0.22 micron syringe filters (Sterlitech,Kent, WA). The cochlea were then micro dissected as described above, apex was perforated and both round and oval windows were punctured. Prepared cochlea were then incubated at 4°C in 4% paraformaldehyde in 0.1 M sodium cacodylate (pH 7.4) overnight with end over end rotation. Cochlea was decalcified in 0.1 M sodium cacodylate (pH 7.4) amended with Immunocal (Decal Chemical Corporation, Congers, NY) for three hours at room temperature. Decalcified bone and tectorial membrane were then removed from the cochlea prior to overnight incubation in 2.5% glutaraldehyde in 0.1 M sodium cacodylate (pH 7.4) overnight at 4°C with agitation. Prepared specimens were then rinsed three times with 0.1 M sodium cacodylate (pH 7.4) for 30 minutes at room temperature prior to 45-minute wash with filtered HPLC grade water. Samples were dried using a Samdri-795 critical point dryer (Tousimis Research Corp, Rockville, MD) following a graded ethanol series to prevent structural damage during imaging. The dried samples were then mounted on aluminum stubs and coated with 15 nm of osmium using a SPI Filgen Osmium Coater (SPI, West Chester, PA). Scanning electron microscope imaging was performed at 3 kV on the JEOL 7900FLV high resolution microscope in the NUANCE Facility at Northwestern University. Images were taken at 2500X and 4500X magnifications for quantification of altered stereocilia morphology.
Western blot analysis
Cochlea extracts were homogenized in Syn-PER buffer (Thermo Scientific, cat. 87793) amended with 1% Triton-X (Sigma-Aldrich, cat. T8787) and 0.5% SDS by Percellys 24 (Bertin, cat. P000669-PR240-A) with three 30 s pulses at 6800 rpm at room temperature. Lysates were then incubated on ice for 10 minutes prior to needle sonication, QSonica Q700 Sonicator (M2 Scientifics, cat Q700), with amplitude set to 20 for 30 s. Lysates were then incubated on ice for 30 minutes, followed by centrifugation for 5 minutes at 10,000 X G at 4°C. Supernatants were collected, and protein concentrations were measured with Pierce BCA protein assay (Thermo Scientific, cat. 23225). Twenty-five micrograms of cochlear lysates were then amended with 6X Loading Buffer and boiled at 100°C for 5 minutes and immediately run on 4%–15% gradient mini Protean TGX gel (Bio-Rad) and transferred to nitrocellulose membrane using Bio-Rad Trans-Blot Turbo transfer system (1.3A –25V-10 minute). Membranes were blocked with 1X Casein blocking solution (Sigma, cat. B6429) for two hours at room temperature and incubated with primary antibody (1:1000) overnight at 4°C. Membranes were washed three times with TBST and incubated with an anti-rabbit IgG-HRP secondary antibody (1:5000, Cell Signal, Cat. 7074S) for an hour at room temperature. Immune reaction on the membranes is captured by incubating with SuperSignal West Femto chemiluminescent substrates (Thermo cat# 34096) and imaged using the Chemidoc imaging system, Bio-Rad. Primary antibodies used for western blot were ARPC2 (Abcam, ab133315), TRIP (Cell signal, cat.13392S) and GAPDH (Abcam, ab181602).
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analyses were performed using Excel (Microsoft, Seattle, WA) and GraphPad Prism. All values in figures with error bars are presented as mean ± SEM. Comparisons of averaged data were performed using the one-way ANOVA for three categories and the paired or unpaired Student’s t test (as indicated) for two categories. Tests with multiple measures at multiple points (i.e., ABR thresholds, DPOAE thresholds, and wave I amplitude) were compared by one-way ANOVA. P values < 0.05 were considered statistically significant. Multiple test correction was performed with the Benjamini-Hochberg correction or FDR strategies as indicated.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| GAPDH | Abcam | Cat# ab181602; RRID:AB_2630358 |
| Vcp | Abcam | Cat# ab111740; RRID:AB_10861709 |
| Psmc5 | Cell signal | Cat# 13392S; RRID:AB_2798204 |
| CtBP2 | BD Transduction Laboratories | Cat# 612044; RRID:AB_399431 |
| Col9a1 | Invitrogen | Cat# PA5-93062; RRID:AB_2806609 |
| GluA2 | Millipore | Cat# MAB397; RRID:AB_2113875 |
| Myosin-VII A | Proteus Bioscience | Cat # 25-6790; RRID:AB_10015251 |
| Neurofilament H | Synaptic Systems | Cat# 171106; RRID:AB_2721078 |
| P4D1 (Ubiquitin antibody) | Santa Cruz | Cat# sc-8017; RRID:AB_2762364 |
| Alexa Fluor 647 Phalloidin | ThermoFisher Scientific | Cat# A22287, RRID:AB_2620155 |
| Uba2 | Cell Signaling | Cat# 5293, RRID:AB_10624868 |
| Arpc2 | Abcam | RRID:AB_133315 |
| Ltbp4 | Abcam | RRID:AB_222844 |
| Hsp90b1 | R&D Systems | RRID:AB7606 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Syn-PER | ThermoFisher Scientific | Cat# 87793 |
| ProteasMax | Promega | Cat# V2071 |
| Sequencing grade modified Trypsin | Promega | Cat# V5111 |
| Lys-C | Promega | Cat# VA1170 |
| Pierce Trypsin Protease | Pierce | Cat# 90058 |
| Casein Blocking Buffer | Sigma | Cat# B6429 |
| SpikeTides TQL - peptides Tpm1 (IQLVEEELD-R*) Col1a1 (ALLLQGSNEIEL-R*), R* = Arg U-13C6;U-15N4 | JPT | N/A (Custom made peptide) |
| Critical Commercial Assays | ||
| TMT10plex Isobaric Mass Tagging Kit | ThermoFisher Scientific | Cat# 90113 |
| TMTpro 16plex Label Reagent Set | ThermoFisher Scientific | Cat# A44521 |
| Pierce BCA Protein Assay Kit | ThermoFisher Scientific | Cat# 23225 |
| RNeasy Plus Mini Kit | QIAGEN | Cat# 74134 |
| Agilent RNA 6000 Nano Kit | Agilent | Cat# 5067-1511 |
| NEBNext® UltraTM RNA Library Prep Kit | New England Bio Labs | Cat# E7770 |
| Tandem Ubiquitin Binding Entities (TUBEs) | LifeSensors | Cat# UM401M |
| PTMScan® Ubiquitin Remnant Motif (K-ε-GG) Kit | Cell Signaling | Cat# 5562 |
| Software and Algorithms | ||
| RStudio | CRAN R Project | Version 1.3.959 |
| Graphpad | Graphpad | Version 8.4.3 |
| STRING | STRING | Version 11.0 |
| PANTHER | PANTHER | Version 15.0 |
| Integrated Proteomics Pipeline(IP2) | Integrated Proteomics Applications, Inc | Version 5.0.1 |
| ProLuCID/SEQUEST algorithm | Yate Laboratory, The Scripps Research Institute | Version 3.1 |
| Census | Yate Laboratory, The Scripps Research Institute | Version 6.0 |
| RawExtract | Yate Laboratory, The Scripps Research Institute | Version 1.9.9 |
| PicoLog system | PCP Piezotronic | Picoscope 2000 series |
| BioSigRP | Tucker Davis Technologies | Version 4.4 |
| iGEAK: Interactive Gene Expression Analysis Kit | Kwangmin Choi | Version: iGEAK v1.0a |
| Other | ||
| DPOAE Speakers | Etymotic | Cat# ER10B+ |
| Speaker compression driver | JBL | Cat# 2446H/J |
| Sound proof chamber | EcKel Audiometric | N/A |
| Genuine grass platinum subdermal needle electrode | Natus Neurology | Cat# F-E2-24 |
| ABR and DPOAE system | Tucker-Davis Technologies | RZ6 Multi I/O Processor |
| Mouse Express (15N, 98%) mouse feed irradiated prepared with spirulina (U-15N, 98%+) | Cambridge Isotope and Harlan Laboratories | Cat# MF-SPIRULINA-N-IR |
| Jupiter C18 resin capillary | Phenomenex | N/A |
| LTQ Velos and Fusion Orbitrap mass spectrometers | Thermo Finnigan | N/A |
| Precellys 24 | Bertin Technology | N/A |
| nanoViper | Thermo Scientific | Cat# 164570 |
| HyperSep SCX Cartridges | ThermoFisher Scientific | Cat# 60108-420 |
| RP IMCSTIPS® | IMCS | Cat# 04T-H6R05-1-20-96 |
| NanoPhotometer® spectrophotometer | IMPLEN | N/A |
| MS raw files | MASSIVE / ProteomeXchange | MSV000084739 / PXD016905 |
| RNA-Seq raw files | Gene Expression Omnibus | GSE160639 |
Highlights.
Hundreds of proteins accumulate in a sound-intensity-dependent manner
Proteasomal subunit mRNA and protein levels elevate after exposure to loud noise
Loud noise induces a burst of cochlear protein ubiquitylation
Ribosomal proteins are upregulated during the process of restoring hearing sensitivity
ACKNOWLEDGMENTS
We thank Mary Ann Cheatham, Margaret Butko, and Ann Hickox for their assistance. This work was supported by R00 DC-013805, W81XWH-19-1-0627, a NU Knowles Hearing Center Pilot award to J.N.S., BX001295 to A.F.R., and T32 MH067564 to M.A.R.
Footnotes
DECLARATION OF INTERESTS
A.F.R. is a co-founder of Otonomy; is a scientific advisory board member; and holds an equity position. UCSD has approved this relationship. Otonomy played no part in this research.
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2020.108431.
REFERENCES
- Bakalarski CE, and Kirkpatrick DS (2016). A Biologist’s Field Guide to Multiplexed Quantitative Proteomics. Mol. Cell. Proteomics 15, 1489–1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balch WE, Morimoto RI, Dillin A, and Kelly JW (2008). Adapting proteostasis for disease intervention. Science 319, 916–919. [DOI] [PubMed] [Google Scholar]
- Butko MT, Savas JN, Friedman B, Delahunty C, Ebner F, Yates JR 3rd, and Tsien RY (2013). In vivo quantitative proteomics of somatosensory cortical synapses shows which protein levels are modulated by sensory deprivation. Proc. Natl. Acad. Sci. USA 110, E726–E735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canzi P, Pecci A, Manfrin M, Rebecchi E, Zaninetti C, Bozzi V, and Benazzo M (2016). Severe to profound deafness may be associated with MYH9-related disease: report of 4 patients. Acta Otorhinolaryngol. Ital 36, 415–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter L, Williams W, Black D, and Bundy A (2014). The leisure-noise dilemma: hearing loss or hearsay? What does the literature tell us? Ear Hear. 35, 491–505. [DOI] [PubMed] [Google Scholar]
- Cheng AG, Cunningham LL, and Rubel EW (2005). Mechanisms of hair cell death and protection. Curr. Opin. Otolaryngol. Head Neck Surg 13, 343–348. [DOI] [PubMed] [Google Scholar]
- Cociorva D, Tabb DL, and Yates JR (2006). Validation of tandem mass spectrometry database search results using DTASelect. Curr. Protoc. Bioinformatics 16, 13.4.1–13.4.14. [DOI] [PubMed] [Google Scholar]
- Elias JE, and Gygi SP (2007). Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods 4, 207–214. [DOI] [PubMed] [Google Scholar]
- Eng JK, McCormack AL, and Yates JR (1994). An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom 5, 976–989. [DOI] [PubMed] [Google Scholar]
- Frost DC, and Li L (2016). High-Throughput Quantitative Proteomics Enabled by Mass Defect-Based 12-Plex DiLeu Isobaric Tags. Methods Mol. Biol 1410, 169–194. [DOI] [PubMed] [Google Scholar]
- Gong TW, Fairfield DA, Fullarton L, Dolan DF, Altschuler RA, Kohrman DC, and Lomax MI (2012). Induction of heat shock proteins by hyperthermia and noise overstimulation in hsf1 −/− mice. J. Assoc. Res. Otolaryngol 13, 29–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouw JW, Krijgsveld J, and Heck AJ (2010). Quantitative proteomics by metabolic labeling of model organisms. Mol. Cell. Proteomics 9, 11–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, Diedrich J, Chu YY, and Yates JR 3rd. (2015). Extracting Accurate Precursor Information for Tandem Mass Spectra by RawConverter. Anal. Chem 87, 11361–11367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Q, Arroyo ED, Smukowski SN, Xu J, Piochon C, Savas JN, Portera-Cailliau C, and Contractor A (2019). Critical period inhibition of NKCC1 rectifies synapse plasticity in the somatosensory cortex and restores adult tactile response maps in fragile X mice. Mol. Psychiatry 24, 1732–1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson D, and Hamernik RP (1995). Biologic bases of noise-induced hearing loss. Occup. Med 10, 513–534. [PubMed] [Google Scholar]
- Hertzano R, and Elkon R (2012). High throughput gene expression analysis of the inner ear. Hear. Res 288, 77–88. [DOI] [PubMed] [Google Scholar]
- Hertzano R, Elkon R, Kurima K, Morrisson A, Chan SL, Sallin M, Biedlingmaier A, Darling DS, Griffith AJ, Eisenman DJ, and Strome SE (2011). Cell type-specific transcriptome analysis reveals a major role for Zeb1 and miR-200b in mouse inner ear morphogenesis. PLoS Genet. 7, e1002309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickox AE, Wong AC, Pak K, Strojny C, Ramirez M, Yates JR 3rd, Ryan AF, and Savas JN (2017). Global Analysis of Protein Expression of Inner Ear Hair Cells. J. Neurosci 37, 1320–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill K, Yuan H, Wang X, and Sha SH (2016). Noise-Induced Loss of Hair Cells and Cochlear Synaptopathy Are Mediated by the Activation of AMPK. J. Neurosci 36, 7497–7510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hjerpe R, Aillet F, Lopitz-Otsoa F, Lang V, England P, and Rodriguez MS (2009). Efficient protection and isolation of ubiquitylated proteins using tandem ubiquitin-binding entities. EMBO Rep. 10, 1250–1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazmierczak M, Harris SL, Kazmierczak P, Shah P, Starovoytov V, Ohlemiller KK, and Schwander M (2015). Progressive Hearing Loss in Mice Carrying a Mutation in Usp53. J. Neurosci 35, 15582–15598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DK, Park Y, Back SA, Kim HL, Park HE, Park KH, Yeo SW, and Park SN (2014). Protective effect of unilateral and bilateral ear plugs on noise-induced hearing loss: functional and morphological evaluation in animal model. Noise Health 16, 149–156. [DOI] [PubMed] [Google Scholar]
- Kimball SR, and Jefferson LS (2012). Induction of REDD1 gene expression in the liver in response to endoplasmic reticulum stress is mediated through a PERK, eIF2α phosphorylation, ATF4-dependent cascade. Biochem. Biophys. Res. Commun 427, 485–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kujawa SG, and Liberman MC (2006). Acceleration of age-related hearing loss by early noise exposure: evidence of a misspent youth. J. Neurosci 26, 2115–2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kujawa SG, and Liberman MC (2009). Adding insult to injury: cochlear nerve degeneration after “temporary” noise-induced hearing loss.J. Neurosci 29, 14077–14085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Prell CG, Yamashita D, Minami SB, Yamasoba T, and Miller JM (2007). Mechanisms of noise-induced hearing loss indicate multiple methods of prevention. Hear. Res 226, 22–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, and Dewey CN (2011). RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12, 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Li Q, Tang J, Xia F, Wu J, and Zeng R (2015). Quantitative Phosphoproteomics Revealed Glucose-Stimulated Responses of Islet Associated with Insulin Secretion. J. Proteome Res 14, 4635–4646. [DOI] [PubMed] [Google Scholar]
- Liberman MC (2016). Noise-Induced Hearing Loss: Permanent Versus Temporary Threshold Shifts and the Effects of Hair Cell Versus Neuronal Degeneration. Adv. Exp. Med. Biol 875, 1–7. [DOI] [PubMed] [Google Scholar]
- Liberman MC (2017). Noise-induced and age-related hearing loss: new perspectives and potential therapies. F1000Res. 6, 927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberman MC, and Dodds LW (1984). Single-neuron labeling and chronic cochlear pathology. III. Stereocilia damage and alterations of threshold tuning curves. Hear. Res 16, 55–74. [DOI] [PubMed] [Google Scholar]
- Lim HH, Jenkins OH, Myers MW, Miller JM, and Altschuler RA (1993). Detection of HSP 72 synthesis after acoustic overstimulation in rat cochlea. Hear. Res 69, 146–150. [DOI] [PubMed] [Google Scholar]
- Lin HW, Furman AC, Kujawa SG, and Liberman MC (2011). Primary neural degeneration in the Guinea pig cochlea after reversible noise-induced threshold shift. J. Assoc. Res. Otolaryngol 12, 605–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Link AJ, Eng J, Schieltz DM, Carmack E, Mize GJ, Morris DR, Garvik BM, and Yates JR 3rd. (1999). Direct analysis of protein complexes using mass spectrometry. Nat. Biotechnol 17, 676–682. [DOI] [PubMed] [Google Scholar]
- Low WK, Kong SW, and Tan MG (2010). Ototoxicity from combined Cisplatin and radiation treatment: an in vitro study. Int. J. Otolaryngol 2010, 523976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Li W, Du X, Ewert DL, West MB, Stewart C, Floyd RA, and Kopke RD (2014). Antioxidants reduce cellular and functional changes induced by intense noise in the inner ear and cochlear nucleus. J. Assoc. Res. Otolaryngol 15, 353–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacCoss MJ, Wu CC, Liu H, Sadygov R, and Yates JR 3rd. (2003). A correlation algorithm for the automated quantitative analysis of shotgun proteomics data. Anal. Chem 75, 6912–6921. [DOI] [PubMed] [Google Scholar]
- Maeda Y, Omichi R, Sugaya A, Kariya S, and Nishizaki K (2017). Cochlear Transcriptome Following Acoustic Trauma and Dexamethasone Administration Identified by a Combination of RNA-seq and DNA Microarray. Otol. Neurotol 38, 1032–1042. [DOI] [PubMed] [Google Scholar]
- Mateo Sánchez S, Freeman SD, Delacroix L, and Malgrange B (2016). The role of post-translational modifications in hearing and deafness. Cell. Mol. Life Sci 73, 3521–3533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maulucci G, Troiani D, Eramo SL, Paciello F, Podda MV, Paludetti G, Papi M, Maiorana A, Palmieri V, De Spirito M, and Fetoni AR (2014). Time evolution of noise induced oxidation in outer hair cells: role of NAD(P)H and plasma membrane fluidity. Biochim. Biophys. Acta 1840, 2192–2202. [DOI] [PubMed] [Google Scholar]
- McAlister GC, Nusinow DP, Jedrychowski MP, Wühr M, Huttlin EL, Erickson BK, Rad R, Haas W, and Gygi SP (2014). MultiNotch MS3 enables accurate, sensitive, and multiplexed detection of differential expression across cancer cell line proteomes. Anal. Chem 86, 7150–7158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald WH, Tabb DL, Sadygov RG, MacCoss MJ, Venable J, Graumann J, Johnson JR, Cociorva D, and Yates JR 3rd. (2004). MS1, MS2, and SQT-three unified, compact, and easily parsed file formats for the storage of shotgun proteomic spectra and identifications. Rapid Commun. Mass Spectrom 18, 2162–2168. [DOI] [PubMed] [Google Scholar]
- Mi H, Muruganujan A, and Thomas PD (2013). PANTHER in 2013: modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Res. 41, D377–D386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H, Poudel S, Muruganujan A, Casagrande JT, and Thomas PD (2016). PANTHER version 10: expanded protein families and functions, and analysis tools. Nucleic Acids Res. 44 (D1), D336–D342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto RI (2008). Proteotoxic stress and inducible chaperone networks in neurodegenerative disease and aging. Genes Dev. 22, 1427–1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto RI (2011). The heat shock response: systems biology of proteotoxic stress in aging and disease. Cold Spring Harb. Symp. Quant. Biol 76, 91–99. [DOI] [PubMed] [Google Scholar]
- Op de Beeck K, Schacht J, and Van Camp G (2011). Apoptosis in acquired and genetic hearing impairment: the programmed death of the hair cell. Hear. Res 281, 18–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paquette ST, Gilels F, and White PM (2016). Noise exposure modulates cochlear inner hair cell ribbon volumes, correlating with changes in auditory measures in the FVB/nJ mouse. Sci. Rep 6, 25056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SK, Venable JD, Xu T, and Yates JR 3rd. (2008). A quantitative analysis software tool for mass spectrometry-based proteomics. Nat. Methods 5, 319–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SK, Aslanian A, McClatchy DB, Han X, Shah H, Singh M, Rauniyar N, Moresco JJ, Pinto AF, Diedrich JK, et al. (2014). Census 2: isobaric labeling data analysis. Bioinformatics 30, 2208–2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, Elias JE, Thoreen CC, Licklider LJ, and Gygi SP (2003a). Evaluation of multidimensional chromatography coupled with tandem mass spectrometry (LC/LC-MS/MS) for large-scale protein analysis: the yeast proteome. J. Proteome Res 2, 43–50. [DOI] [PubMed] [Google Scholar]
- Peng J, Schwartz D, Elias JE, Thoreen CC, Cheng D, Marsischky G, Roelofs J, Finley D, and Gygi SP (2003b). A proteomics approach to understanding protein ubiquitination. Nat. Biotechnol 21, 921–926. [DOI] [PubMed] [Google Scholar]
- Rabinowitz PM (2000). Noise-induced hearing loss. Am. Fam. Physician 61, 2749–2756, 2759–2760. [PubMed] [Google Scholar]
- Rauniyar N, and Yates JR 3rd. (2014). Isobaric labeling-based relative quantification in shotgun proteomics. J. Proteome Res 13, 5293–5309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regard JB, Scheek S, Borbiev T, Lanahan AA, Schneider A, Demetriades AM, Hiemisch H, Barnes CA, Verin AD, and Worley PF (2004). Verge: a novel vascular early response gene. J. Neurosci 24, 4092–4103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson D (1983). Functional significance of dendritic swelling after loud sounds in the guinea pig cochlea. Hear. Res 9, 263–278. [DOI] [PubMed] [Google Scholar]
- Ruel J, Wang J, Rebillard G, Eybalin M, Lloyd R, Pujol R, and Puel JL (2007). Physiology, pharmacology and plasticity at the inner hair cell synaptic complex. Hear. Res 227, 19–27. [DOI] [PubMed] [Google Scholar]
- Ryan AF, Kujawa SG, Hammill T, Le Prell C, and Kil J (2016). Temporary and Permanent Noise-induced Threshold Shifts: A Review of Basic and Clinical Observations. Otol. Neurotol 37, e271–e275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago AM, Gonçalves DL, and Morano KA (2020). Mechanisms of sensing and response to proteotoxic stress. Exp. Cell Res 395, 112240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders GH, and Griest SE (2009). Hearing loss in veterans and the need for hearing loss prevention programs. Noise Health 11, 14–21. [DOI] [PubMed] [Google Scholar]
- Savas JN, Ribeiro LF, Wierda KD, Wright R, DeNardo-Wilke LA, Rice HC, Chamma I, Wang YZ, Zemla R, Lavallée-Adam M, et al. (2015). The Sorting Receptor SorCS1 Regulates Trafficking of Neurexin and AMPA Receptors. Neuron 87, 764–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savas JN, Wang YZ, DeNardo LA, Martinez-Bartolome S, McClatchy DB, Hark TJ, Shanks NF, Cozzolino KA, Lavallée-Adam M, Smukowski SN, et al. (2017). Amyloid Accumulation Drives Proteome-wide Alterations in Mouse Models of Alzheimer’s Disease-like Pathology. Cell Rep. 21, 2614–2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiavon E, Smalley JL, Newton S, Greig NH, and Forsythe ID (2018). Neuroinflammation and ER-stress are key mechanisms of acute bilirubin toxicity and hearing loss in a mouse model. PLoS ONE 13, e0201022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt H, Roemer A, Zeilinger C, Salcher R, Durisin M, Staecker H, Lenarz T, and Warnecke A (2018). Heat Shock Proteins in Human Perilymph: Implications for Cochlear Implantation. Otol. Neurotol 39, 37–44. [DOI] [PubMed] [Google Scholar]
- Schubert OT, Röst HL, Collins BC, Rosenberger G, and Aebersold R (2017). Quantitative proteomics: challenges and opportunities in basic and applied research. Nat. Protoc 12, 1289–1294. [DOI] [PubMed] [Google Scholar]
- Shi L, Chang Y, Li X, Aiken S, Liu L, and Wang J (2016). Cochlear Synaptopathy and Noise-Induced Hidden Hearing Loss. Neural Plast. 2016, 6143164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strimbu CE, Prasad S, Hakizimana P, and Fridberger A (2019). Control of hearing sensitivity by tectorial membrane calcium. Proc. Natl. Acad. Sci. USA 116, 5756–5764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M, Santos A, Doncheva NT, Roth A, Bork P, et al. (2017). The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 45 (D1), D362–D368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabb DL, McDonald WH, and Yates JR 3rd. (2002). DTASelect and Contrast: tools for assembling and comparing protein identifications from shotgun proteomics. J. Proteome Res 1, 21–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas PD, Kejariwal A, Guo N, Mi H, Campbell MJ, Muruganujan A, and Lazareva-Ulitsky B (2006). Applications for protein sequence-function evolution data: mRNA/protein expression analysis and coding SNP scoring tools. Nucleic Acids Res. 34, W645–W650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washburn MP, Wolters D, and Yates JR 3rd. (2001). Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat. Biotechnol 19, 242–247. [DOI] [PubMed] [Google Scholar]
- Weekes MP, Tomasec P, Huttlin EL, Fielding CA, Nusinow D, Stanton RJ, Wang ECY, Aicheler R, Murrell I, Wilkinson GWG, et al. (2014). Quantitative temporal viromics: an approach to investigate host-pathogen interaction. Cell 157, 1460–1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G, Paige JS, and Jaffrey SR (2010). Global analysis of lysine ubiquitination by ubiquitin remnant immunoaffinity profiling. Nat. Biotechnol 28, 868–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T, Park SK, Venable JD, Wohlschlegel JA, Diedrich JK, Cociorva D, Lu B, Liao L, Hewel J, Han X, et al. (2015). ProLuCID: An improved SEQUEST-like algorithm with enhanced sensitivity and specificity. J. Proteomics 129, 16–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Cai Q, Bard J, Jamison J, Wang J, Yang W, and Hu BH (2015). Variation analysis of transcriptome changes reveals cochlear genes and their associated functions in cochlear susceptibility to acoustic overstimulation. Hear. Res 330 (Pt A), 78–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Cai Q, Vethanayagam RR, Wang J, Yang W, and Hu BH (2016). Immune defense is the primary function associated with the differentially expressed genes in the cochlea following acoustic trauma. Hear. Res 333, 283–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida N, Kristiansen A, and Liberman MC (1999). Heat stress and protection from permanent acoustic injury in mice. J. Neurosci 19, 10116–10124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Fenyö D, and Neubert TA (2009). Evaluation of the variation in sample preparation for comparative proteomics using stable isotope labeling by amino acids in cell culture. J. Proteome Res 8, 1285–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession number for the MS data reported in this paper is MASSIVE: MSV000084739 and ProteomeXchange: PXD016905. The accession number for the RNA-seq data reported in this paper is GEO: GSE160639.


