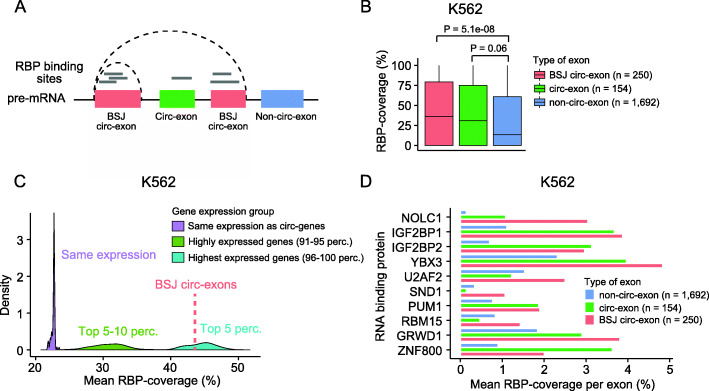
Fig. 3.
Exons comprising circRNAs are enriched with RBP binding sites. a Illustration of exon categories in genes forming circRNAs. Backsplice junction circularizing exons (BSJ circ-exons, red) are involved in the backsplicing event. Circularizing exons (circ-exons, green) are potentially part of circRNAs, if not spliced out. Non-circularizing exons (non-circ-exons, blue) are not part of circRNAs. Gray lines represent RBP binding sites. To evaluate the fraction of exons covered by RBP binding sites overall, the individual overlapping RBP binding sites were merged. b RBP binding site coverage for each category of exons in genes that produce highly expressed (top 1%) circRNAs in K562 (n = 124). P values obtained by Wilcoxon rank sum test. c Comparison of RBP coverage between BSJ circ-exons from top 1% circRNAs and exons in groups of genes of different expression levels (K562). The mean RBP coverage of the highly expressed BSJ circ-exons (n = 250) is 44% (red punctuated line; median = 36%). Exons randomly sampled from genes while ensuring the same expression profile as genes producing highly expressed circRNAs (circ-genes) have a much lower mean coverage of 23% (purple; median = 0%). Exons of highly expressed genes (top 5–10 percentiles) also showed a lower mean coverage of 31% (green; median = 3%), while the most highly expressed genes (top 5 percentiles) had a slightly higher mean of 45% (blue; median = 43%). The random sampling procedures were repeated with 100 iterations. Empirical P values for BSJ circ-exons vs. same expression, P < 0.01; BSJ circ-exons vs. Top5–10, P < 0.01; BSJ circ-exons vs Top5, P = 0.67. d Mean RBP coverage per exon for individual RBPs (K562). All RBPs shown here have significantly more target sites in highly expressed BSJ circ-exons than in non-circ-exons of the same genes (FDR < 0.1, Wilcoxon rank sum rest). Only RBPs with at least 20 distinct binding sites in total are considered