Figure 2.
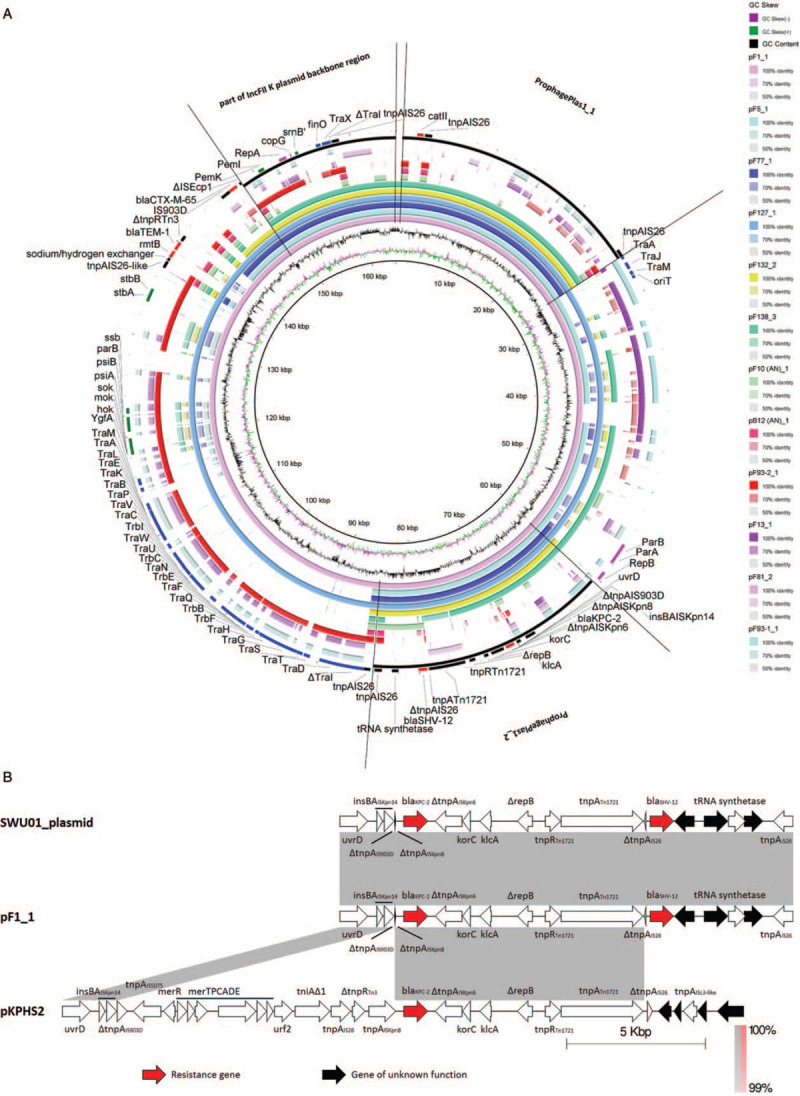
(A) Alignment of the plasmid sequences of the K. pneumoniae strains. BLASTN-based whole genome comparison was performed and visualized using BRIG to exhibit the architecture and gene repertoire of a total of twelve K. pneumoniae plasmids, using the plasmid sequence of pF1_1 as a reference. Part of the IncFII plasmid backbone region, ProphagePlas1_1, ProphagePlas1_2, and the significant genes are indicated by rectangles. (B) Transposon carrying blaKPC-2 located in our isolates (ΔΔTn1721-blaKPC-2) was compared with that in pKPHS2 (ΔTn1721- blaKPC-2) and that in the unnamed plasmid of SWU01 (ΔΔTn1721-blaKPC-2). Regions of synteny between adjacent schematics are indicated by the shaded areas; the matching percentage nucleotide sequence identity for each such region is indicated. These schematics are drawn to scale. K. pneumoniae: Klebsiella pneumoniae.
