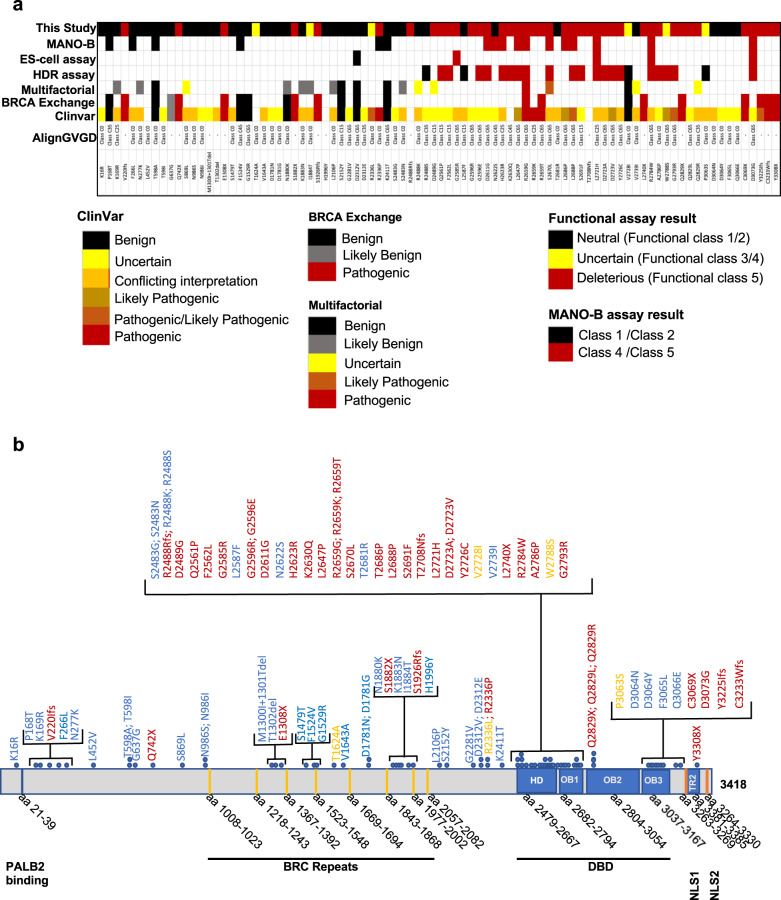
Fig. 6. Classification of variants analyzed using the VarCall method.
a Variants analyzed using VarCall method are compared to ClinVar, Align GVGD, BRCA Exchange, classification by multifactorial likelihood ratio and published BRCA2 functional assays. Colored boxes represent different classifications listed below. The MANO-B (mixed-allnominated-in-one (MANO)-BRCA) method of classification includes the classification by Ikegami et al. (2020), the ES assay includes the classification of variants using mouse ES cell-based assay reported by Mesman et al. (2019) and the HDR assay includes the classification of variants using HR assay reported by Guidugli et al. (2018)18,19,33. The Multifactorial classifications are based on the study by Parson et al.51. b Schematic representation of classified variants in different domains of BRCA2. Neutral, hypomorphic and pathogenic variants are marked in blue, yellow, and magenta colors, respectively. Different domains are marked below with the range of amino acids (aa) containing in each domain.