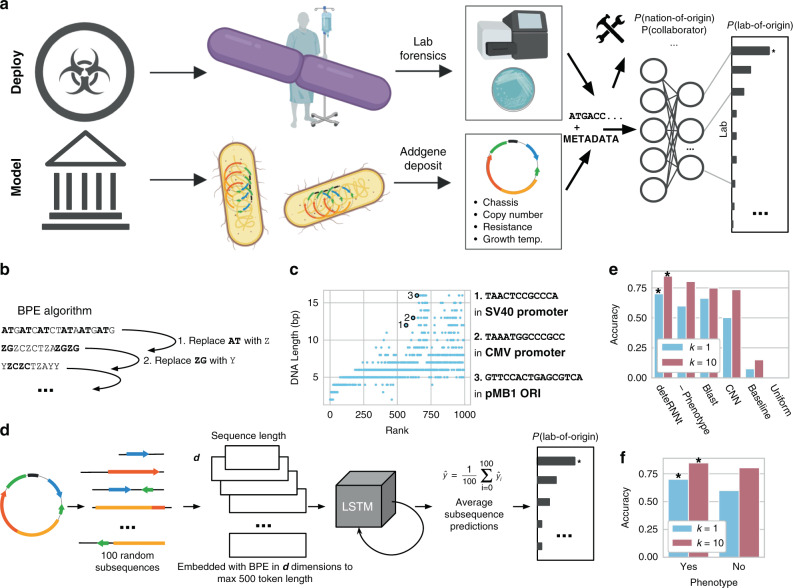
Fig. 1. Deep learning on DNA motifs and minimal phenotype information enables state-of-the-art genetic engineering attribution.
a The Addgene plasmid repository (bottom) provides a model through which to study the deployment scenario (top) for genetic engineering attribution. In the model scenario, research laboratories engineer organisms and share their genetic designs with the research community by depositing the DNA sequence and phenotypic metadata information to Addgene. In the corresponding deployment scenario, a genetically engineered organism of unknown origin is obtained, for example, from an environmental sample, lab accident, misuse incident, or case of disputed authorship. By characterizing this sample in the laboratory with sequencing and phenotype experiments, the investigator identifies the engineered sequence and phenotype information. In either case, the sequence and phenotype information are input to an attribution model which predicts the probability the organism originated from individuals connected to a set of known labs, enabling further conventional investigation; * indicates a hypothetical “best match” predicted by the imagined model. Above, the same information may be input to a wider toolkit of methods which provide actionable leads and characterization of the sample to support the investigator. b DNA motifs are inferred through the Byte Pair Encoding (BPE) algorithm25, which successively merges the most frequently occurring pairs of tokens to compress input sequences into a vocabulary larger than the traditional four DNA bases. Progressively, sequences become shorter and new motif tokens become longer. c BPE on the training set of Addgene plasmid sequences. The x axis shows the tokens rank-ordered by frequency in the sequence set (decreasing). The y axis shows token length, in base pairs. Example tokens (bold, numbered) are linked to biologically meaningful sequence motifs. d The deteRNNt method takes 100 random subsequences from the plasmid encoded with BPE and embeds them into a continuous space via a learned word embedding59 matrix layer. These (potentially variable-length) sequences are processed by an LSTM network. We average the predictions from each subsequence to obtain a softmax probability that the plasmid originated in a given lab. e Top-k prediction accuracy on the test set. Compared: deteRNNt, deteRNNt trained without phenotype, BLASTn, CNN deep-learning state-of-the-art method21, a baseline guessing the most abundant labs from the training set, and guessing uniformly randomly (so low, cannot be seen).* indicates P < 10−10, by Welch’s two-tailed t test on n = 30 × 50% bootstrap replicates compared to BLAST. f Top-k prediction accuracy on test set with and without phenotype information. * indicates P < 10−10, by Welch’s two-tailed t test on n = 30 × 50% bootstrap replicates compared to no phenotype.