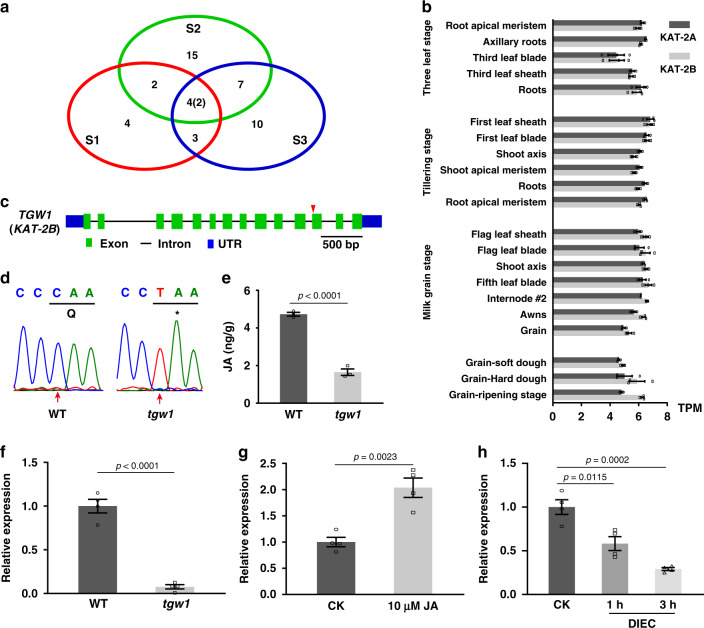
Fig. 2. Molecular cloning of TGW1.
a Venn diagram to show distributions of homozygous SNPs in three groups of homozygous BC1F2 mutants. S1 to S3 represented three groups of homozygous BC1F2 mutants. Each number represents numbers of homozygous SNPs. Only two nonsynonymous homozygous SNPs presented in three groups. b The expression of KAT-2B (TraesCS6B01G432600) and KAT-2A (TraesCS6A01G392400) in different tissues at various developmental stages. n = 3 independent replicates. c Genomic structure of TGW1 (KAT-2B) gene. The red triangle points to the position of the point mutation in tgw1. d Verification of the SNP in the KAT-2B (TGW1) gene in WT and tgw1 by sequencing. e Contents of JA in leaves of WT and tgw1. n = 3. f Relative expression level of Cell Number Regulator 6 (CNR6) in WT and tgw1 mutant. n = 4. g The expression level of CNR6 upon 10 μM JA treatments in WT. n = 4. h The expression level of CNR6 upon 200 μM JA inhibitor DIEC treatments in WT. n = 4. Data are represented as mean ± SEM in b, and e–h. p values are indicated by two-tailed unpaired t test in e–h. Source data underlying Figs. 2a, b, e–h are provided as a Source Data file.