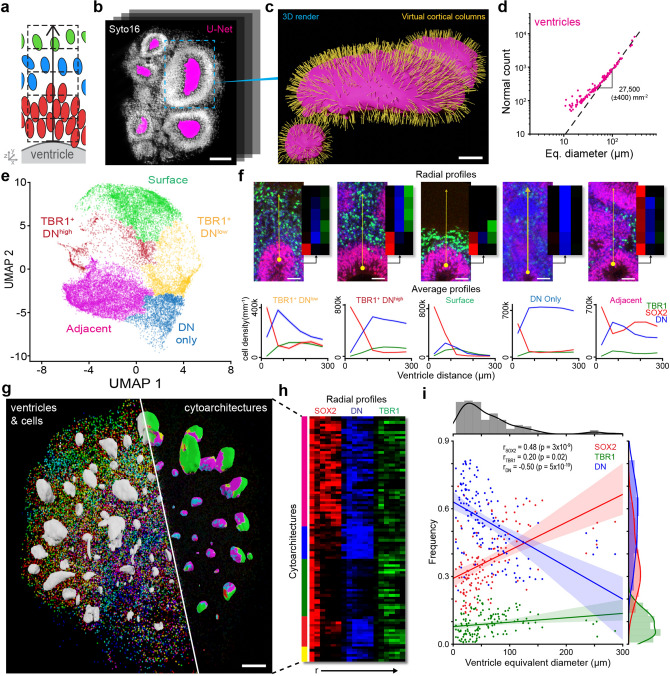
Figure 3.
SCOUT analysis of regional architectures. (a) Scheme of automated cytoarchitecture analysis. We quantified radial organization of cell populations around ventricles using “virtual cortical columns” 50 µm in diameter and 300 µm high, perpendicular to the ventricle surface. (b) Demonstration of automated ventricle segmentation using U-Net convolutional neural network. Representative optical section of a volumetric dataset with detected ventricles in magenta. (c) A 3D render of ventricle highlighted in panel B with normals used to orient virtual cortical columns shown in yellow. (d) Graph showing that the total number of normals per ventricle depends on the ventricle’s surface area. (e) UMAP embedding of detected cytoarchitectures in a single organoid color-coded according to each cluster. (f) Representative image and average profile plot of individual cytoarchitecture clusters showing the radial distribution of SOX2 (red), double negatives (blue) and TBR1 (green) cells. Scale bar, 50 µm (g) 3D render of segmented cells and ventricles from a day 35 organoid. On the left side ventricles are white and six cell populations are colored according to the index in Fig. 2l: SOX2 in red, SOX2-adjacent in magenta, co-adjacent in yellow, TBR1-adjacent in cyan, TBR1 in green and core DN in blue. On the right, we mapped the detected cytoarchitectures on the surface of rendered ventricles using the colors in (f). Scale bar = 200 µm (h) Three-channel heat map from 100 random cytoarchitectures. Each row shows the number of cells detected in all six 50 µm increments moving away from the ventricle surface. Intensity of red, blue and green represent SOX2, DN and TBR1, respectively. (i) The frequency of SOX2, DN and TBR1 cells detected in a ventricle’s virtual cortical columns correlates with the ventricle equivalent diameter. Strongest correlation occurs for decreased DN and increased SOX2 in larger ventricles.