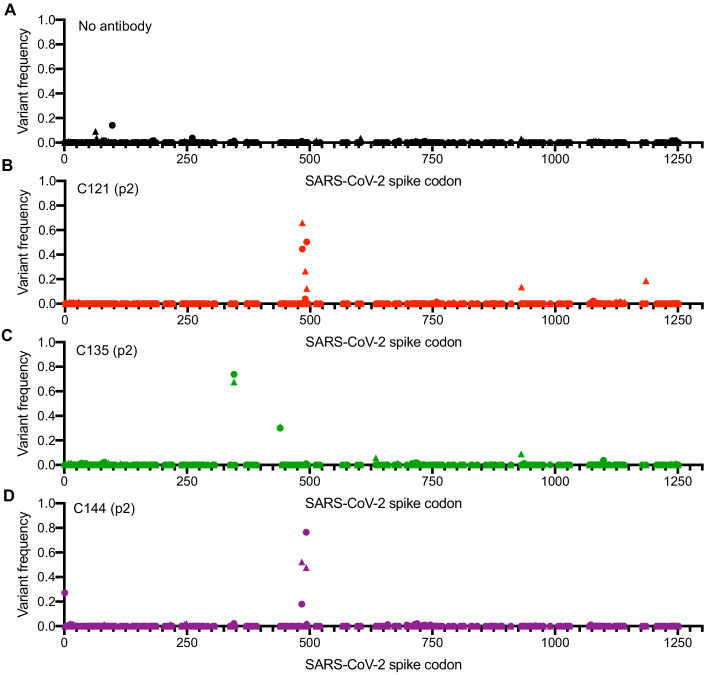
Figure 2. Analysis of S-encoding sequences following rVSV/SARS-CoV-2/GFP replication in the presence of neutralizing monoclonal antibodies.
(A–D) Graphs depict the S codon position (X-axis) and the frequency of non-synonymous substitutions (Y-axis) following the second passage (p2) of rVSV/SARS-CoV-2/GFP on 293T/ACE2(B) cells in the absence of antibody or plasma (A), or in the presence of 10 μg/ml C121 (B), C135 (C) or C144 (D). Results are shown for both rVSV/SARS-CoV-2/GFP variants (One replicate each for rVSV/SARS-CoV-2/GFP1D7 and rVSV/SARS-CoV-2/GFP2E1 - the frequency of 1D7 mutations is plotted as circles and 2E1 mutations as triangles).