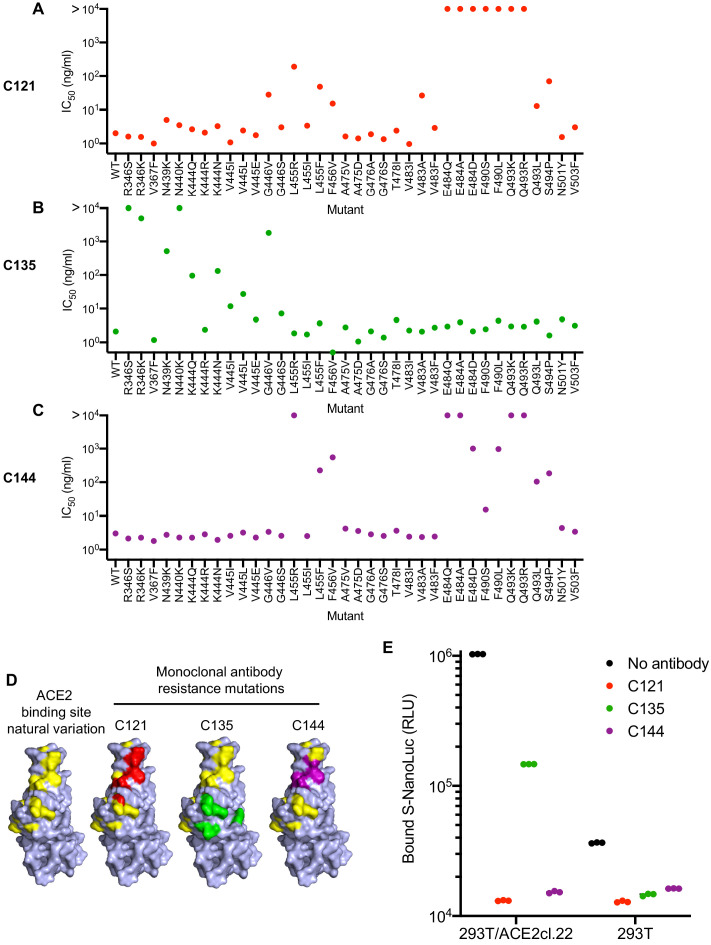
Figure 7. Effects of naturally occurring RBD amino-acid substitutions on S sensitivity to neutralizing monoclonal antibodies.
(A–C) Neutralization of HIV-based reporter viruses pseudotyped with SARS-CoV-2 S proteins harboring the indicated naturally occurring substitutions. 293T/ACE2cl.22 cells were inoculated with equivalent doses of each pseudotyped virus in the presence of increasing amount of C121 (A) C135 (B) or C144 (C). Mean IC50 values were calculated for each virus-antibody combination from two independent experiments. (D) Position of substitutions conferring neutralization resistance relative to the amino acids close to the ACE2 binding site whose identity varies in global SARS-CoV-2 sequences. The RBD structure (from PDB 6M17 Yan et al., 2020) is depicted with naturally varying amino acids close to the ACE2 binding site colored in yellow. Amino acids whose substitution confers partial or complete (IC50 > 10 μg/ml) resistance to each monoclonal antibody in the HIV-pseudotype assays are indicated for C121 (red) C135 (green) and C144 (purple). (E) Binding of S-NanoLuc fusion protein in relative light units (RLU) to 293T or 293T/ACE2cl.22 cells after preincubation in the absence or presence of C121, C135, and C144 monoclonal antibodies. Each symbol represents a technical replicate.