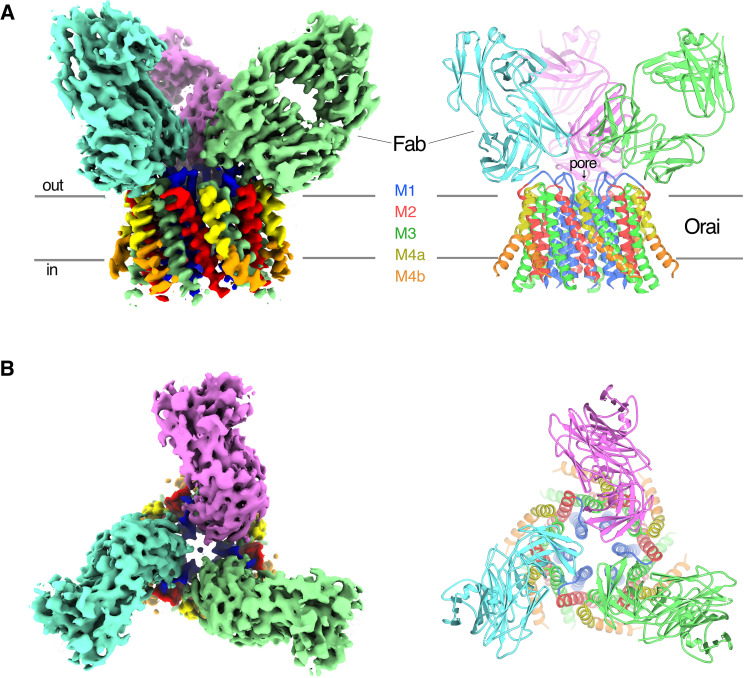
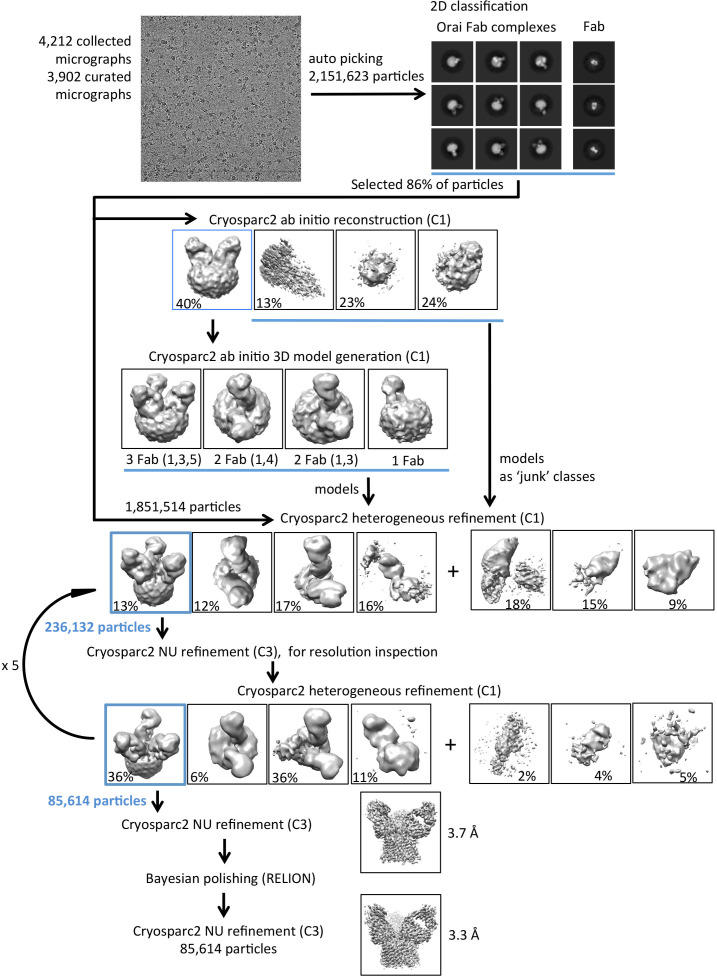
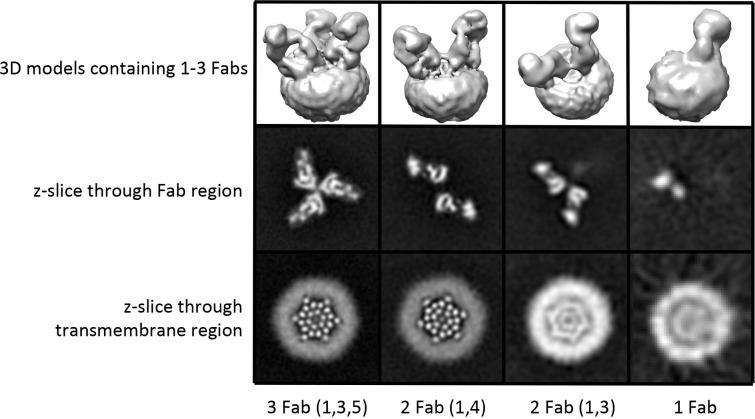
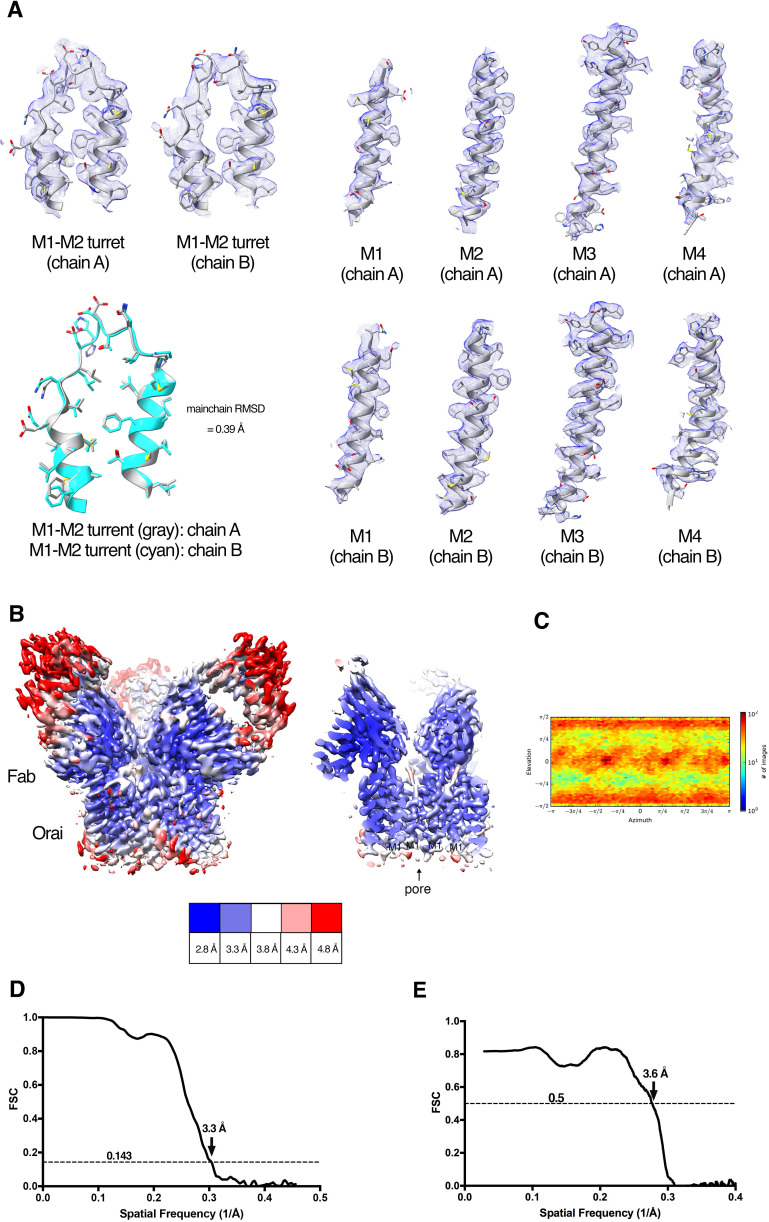
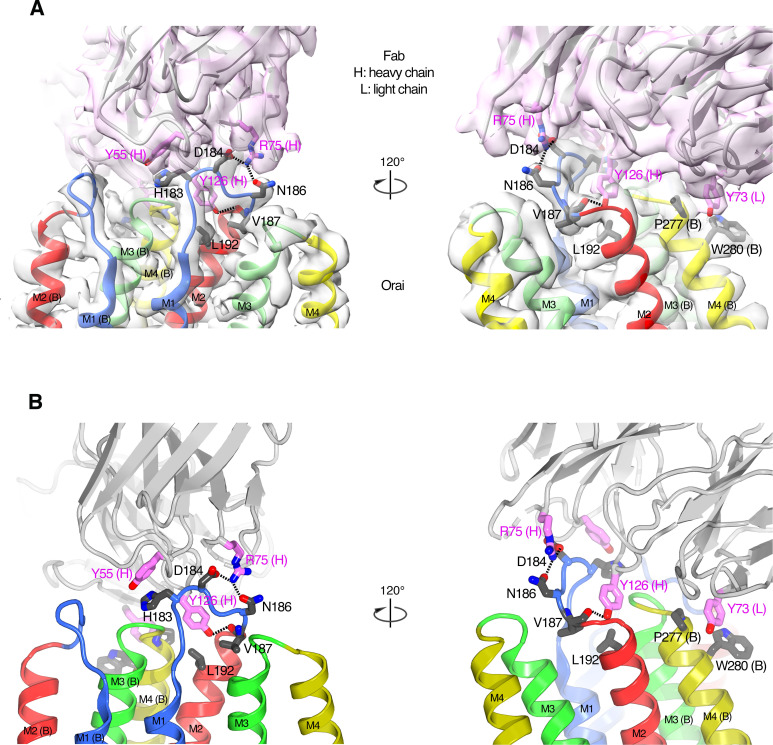
Figure 2. OraiH206A–Fab complex.
(A and B) Overall structure of the complex is shown from the perspective of the membrane (A) and from the extracellular side (B). The cryo-EM map (left) and the cartoon representation of the structure (right) are shown. Each Fab is colored individually; the α-helices of Orai are colored as indicated. Horizontal bars in (A) denote approximate boundaries of the plasma membrane.