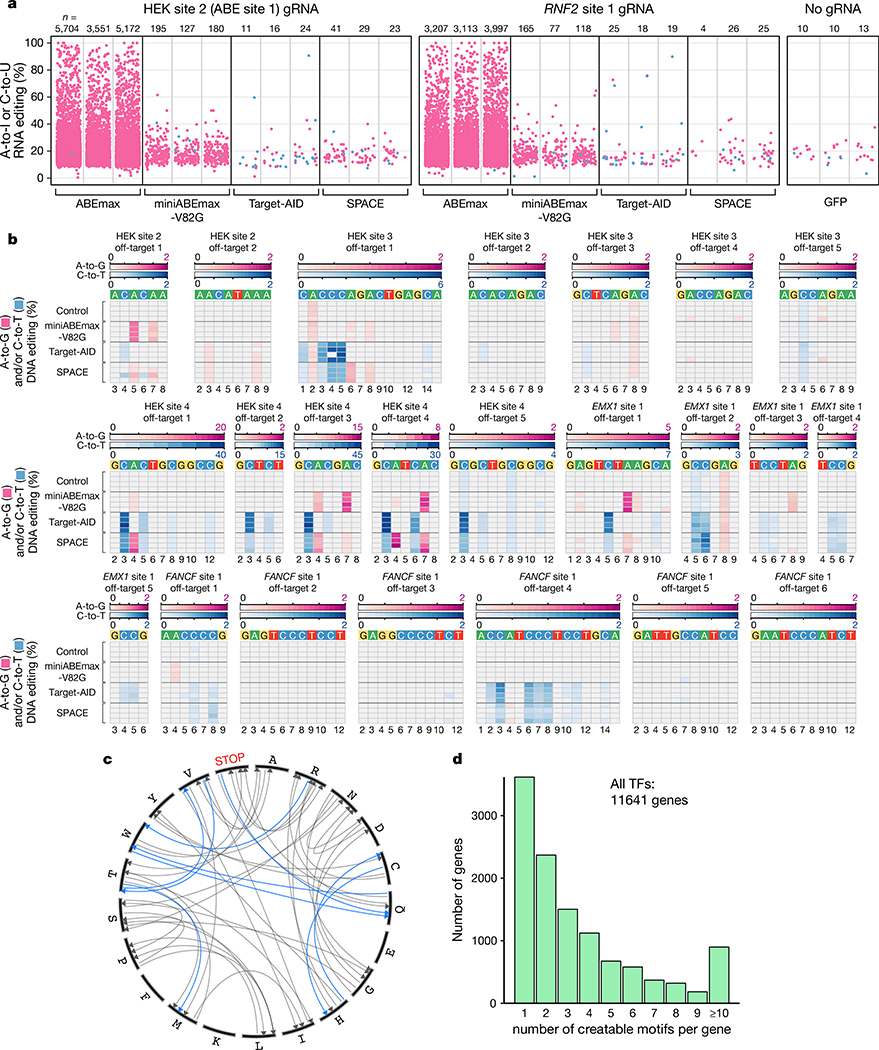
Figure 2. RNA and DNA off-target editing, potential amino acid modifications, and number of genes with potential transcription factor binding sites induced by SPACE.
a, Jitter plots show transcriptomic A-to-I (pink) and C-to-U (blue) mutations detected in RNA-seq experiments from HEK293T cells in which ABEmax, miniABEmax-V82G, Target-AID, or SPACE were co-expressed with either a HEK site 2 or RNF2 site 1 gRNA. GFP control cells express no gRNA. Data are shown from three independent replicates. n= number of combined adenines and cytosines modified. b, Heat maps showing A-to-G (pink) and C-to-T (blue) DNA off-target editing frequencies of nCas9 (Control), miniABEmax-V82G, Target-AID, or SPACE co-expressed in HEK293T cells with gRNAs targeted to HEK sites 2–4, EMX1 site 1, or FANCF site 1 (n= 4 independent replicates). The respective off-target editing windows for each site are shown. Location in the protospacer is indicated at the bottom with 1 being the most PAM-distal position. c, Circos plot showing amino acid changes that can be induced by SPACE (gray), including amino acid changes in specific codon context, that are uniquely enabled (blue). d, Bar plots showing computationally determined number of genes (y-axis) that could be targeted by SPACE to install 1–10 (or more) transcription factor (TF) binding sites (x-axis) in proximity of the transcription start site of coding genes in the human genome. Sites were filtered to contain a preferential SPACE editing window (C3-C4-A5 with 1 being the most PAM-distal position) and a canonical NGG PAM (Online Methods).