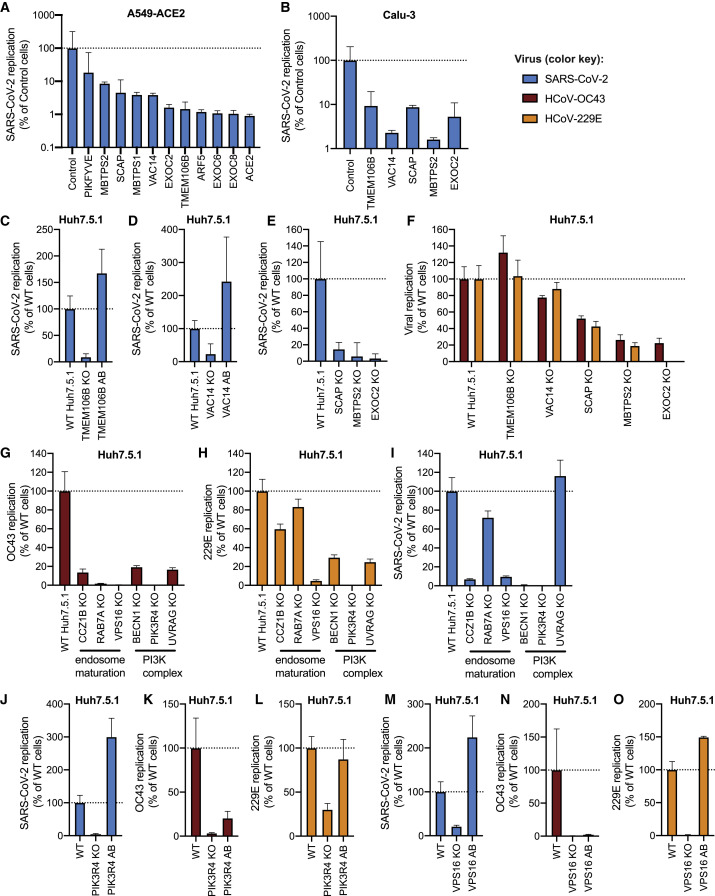
Figure 3.
KO of Candidate Host Factor Genes Reduces Coronavirus Infection
(A) Quantitative reverse-transcriptase PCR (RT-qPCR) quantification of intracellular SARS-CoV-2 levels in RNP-edited A549-ACE2 cells. A non-targeting sgRNA was used as control. Cells were infected using MOI = 0.1 and harvested at 72 h post-infection (hpi).
(B) RT-qPCR quantification of intracellular SARS-CoV-2 levels in Calu-3 cells lentivirally transduced with Cas9/sgRNA cassettes targeting the indicated genes. A non-targeting sgRNA was used as control. Cells were infected using MOI = 0.1 and harvested at 48 hpi.
(C) RT-qPCR quantification of intracellular SARS-CoV-2 levels in WT Huh7.5.1, TMEM106B KO, or TMEM106B KO cells with TMEM106B cDNA add-back (AB). Cells were infected using MOI = 0.1 and harvested at 24 hpi.
(D) RT-qPCR quantification of intracellular SARS-CoV-2 levels in WT Huh7.5.1, VAC14 KO, or VAC14 KO cells with VAC14 cDNA AB. Cells were infected using MOI = 0.1 and harvested at 24 hpi.
(E) RT-qPCR quantification of intracellular SARS-CoV-2 levels in WT Huh7.5.1, SCAP KO, MBTPS2 KO, or EXOC2 KO cells. Cells were infected using MOI = 0.1 and harvested at 24 hpi.
(F) RT-qPCR quantification of intracellular OC43 and 229E RNA levels in WT and TMEM106B, VAC14, SCAP, MBTPS2, or EXOC2 KO Huh7.5.1 cells. Cells were infected using MOI = 0.05 (229E) and MOI = 3 (OC43) and harvested at 48 hpi.
(G–I) RT-qPCR quantification of intracellular viral RNA for (G) OC43, (H) 229E, or (I) SARS-CoV-2 in WT Huh7.5.1 cells or cell lines deficient in CCZ1B, RAB7A, VPS16, BECN1, PIK3R4, or UVRAG.
(J–L) RT-qPCR quantification of intracellular viral RNA for (J) SARS-CoV-2, (K) OC43, or (L) 229E in WT, PIK3R4 KO, or PIK3R4 KO cells with PIK3R4 cDNA AB.
(M–O) RT-qPCR quantification of intracellular viral RNA for (M) SARS-CoV-2, (N) OC43, or (O) 229E in WT, VPS16 KO, or VPS16 KO cells with VPS16 cDNA AB.
For SARS-CoV-2 infection, viral N gene transcripts were normalized to cellular RNaseP. For OC43 and 229E experiments, viral RNA was normalized to 18S RNA. For all RT-qPCR experiments, results are displayed relative to infection in WT cells, and data represent means ± SEM from three biological samples.