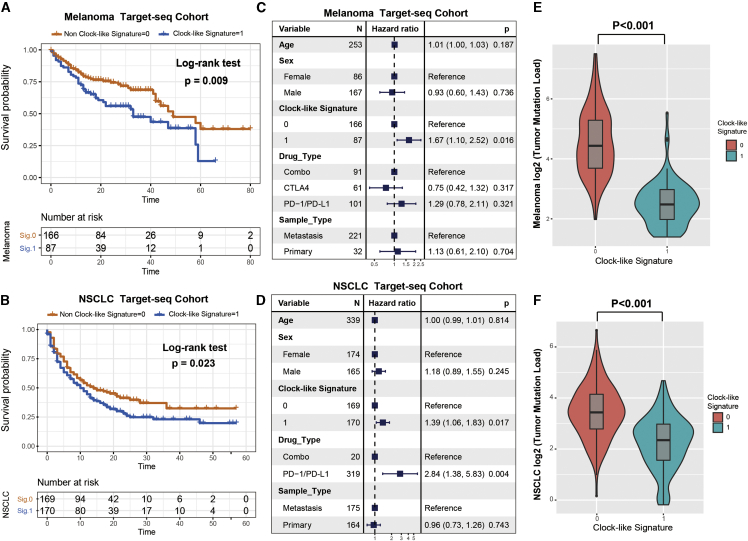
Figure 5.
Clock-like mutational signature extracted from the targeted NGS-panel sequencing datasets also associated with immune non-responding
SigMA analysis tool was utilized to decompose the mutational signatures. (A and B) Kaplan-Meier survival analysis was utilized to analyze clock-like mutational signature grouping in t-melanoma and t-NSCLC cohorts. p value was calculated by log-rank test. (C and D) Forest plot representation of the multivariate Cox regression model with adjustment for confounding clinical factors. p value was inferred by multivariate Cox regression model. (E and F) Tumor mutation load was assessed by clock-like mutation signature status in t-melanoma and t-NSCLC. p value was calculated by Wilcoxon rank-sum test. Error bars represented the 95% confidence interval.