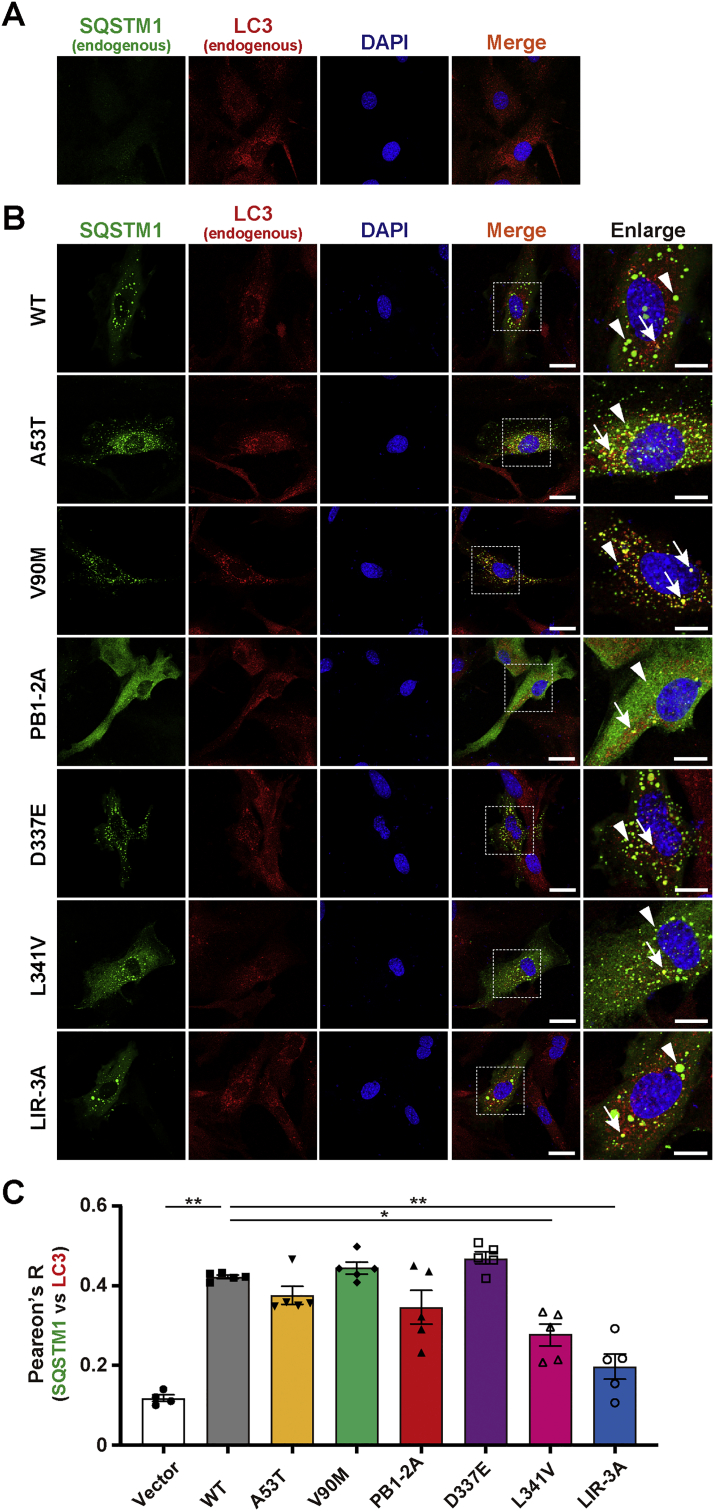
Fig. 1.
Subcellular distribution of SQSTM1 mutants in Sqstm1 deficient mouse embryonic fibroblast. Sqstm1 deficient mouse embryonic fibroblasts (Sqstm1-KO MEFs) were transfected with FLAG-tagged pCIneo_SQSTM1 constructs. (A) Representative images for the control experiment (Vector). (B) Representative images for SQSTM1_WT (WT), PB1-domain ALS-linked variants (A53T and V90M), PB1-domain artificial mutant (PB1-2A), LIR ALS-linked variants (D337E and L341V) and LIR artificial mutant (LIR-3A)-expressing cells. Images for SQSTM1 and LC3 were obtained by staining with anti-SQSTM1 (p62-C) and anti-LC3 antibodies, respectively. Nuclei were counterstained with DAPI. High-resolution magnified images are shown on the right. Arrows and arrowheads indicate LC3-positive and -negative SQSTM1-bodies, respectively. Scale bars for merged and enlarged images indicate 20 μm and 10 μm, respectively. (C) Pearson's correlation coefficient analysis of the colocalization between SQSTM1 and LC3. Values of Pearson's R are expressed as mean ± S.E.M. (n = 5 biological replicates). Individual data points are also shown. Statistical significances were evaluated by one-way ANOVA with Tukey's multiple comparison test. The results of comparisons between WT and other conditions are only shown (*p < 0.05, **p < 0.01).