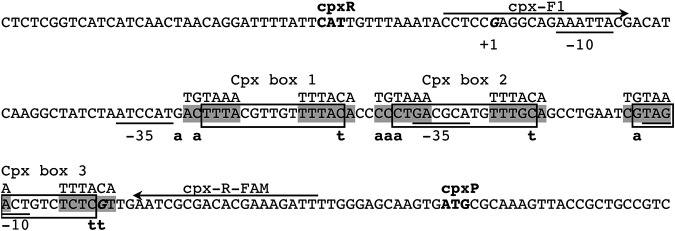
FIG 7.
Predicted regulatory elements in the intergenic region of divergently transcribed cpxR and cpxP. The locations of CpxR binding sites are boxed, and point mutations made to predicted TyrR binding sites (shaded) overlapping each are indicated below the sequence. The consensus sequence for the TyrR binding site is shown above the predicted sites. The cpxR and cpxP start codons are in bold, transcription start sites (+1) are in bold italics, and the −35 and −10 elements of the sigma-70 promoters are underlined. The primer binding sites used to generate probes for EMSAs are marked with arrows. The locations of promoter elements and CpxR binding sites are inferred from orthologous sequences in the E. coli cpx promoters, which share a high degree of sequence conservation (see Fig. S7 in the supplemental material).