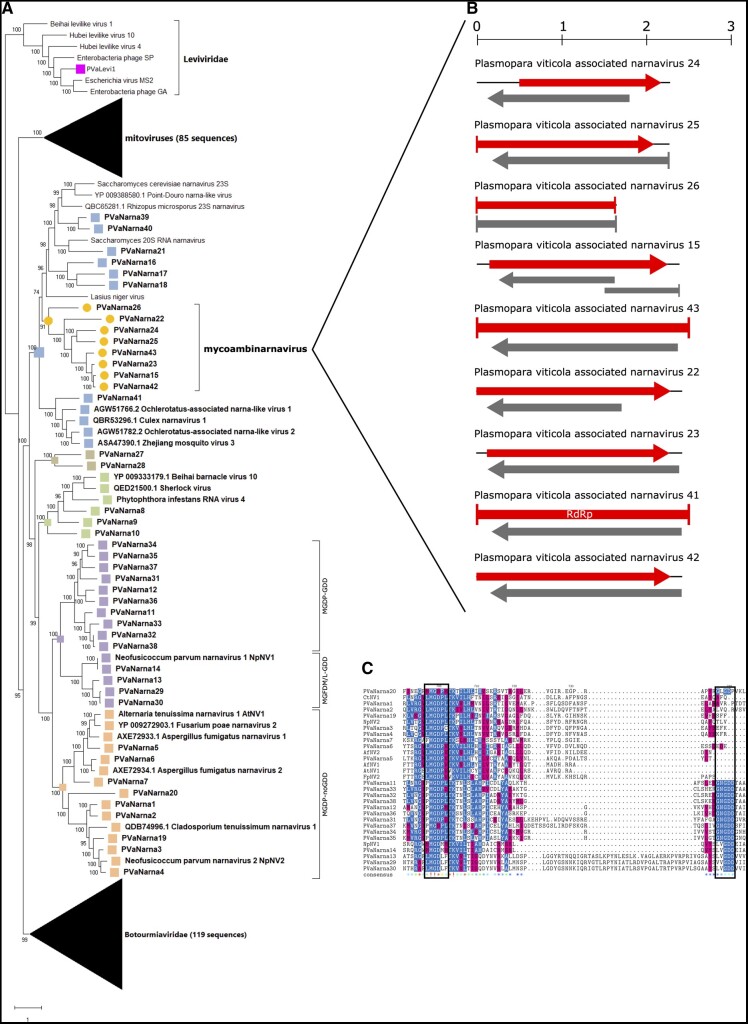
Figure 1.
(A) Lenarviricota phylogenetic tree computed by IQ-TREE stochastic algorithm to infer phylogenetic trees by maximum likelihood. Model of substitution: VT+F + I+G4. Consensus tree is constructed from 1,000 bootstrap trees. Log-likelihood of consensus tree: −377870.722. At nodes the percentage bootstrap values. The tree focus is on narnaviruses. Red stars indicate viruses with missing GDD motif. Color dots correspond to established or proposed new genera. (B) Genomic organization of nine ambinarnaviruses with a second ORF in reverse genomic orientation. Top ruler indicates the length in kb. (C) Alignment of narna-like viruses without the GDD domain and some of the closest homologues with the GDD domain. In the consensus row an exclamation mark ‘!’ stands for a conserved amino acid, while an asterisk ‘*’ stands for positions in which there is a majority of sequences agreeing. Positions in which the sequences disagree are left blank in the consensus sequence. Color code indicates the aminoacidic properties. The two boxes show the GDD and the conserved MGDP/M or L conserved motifs.