Figure 2.
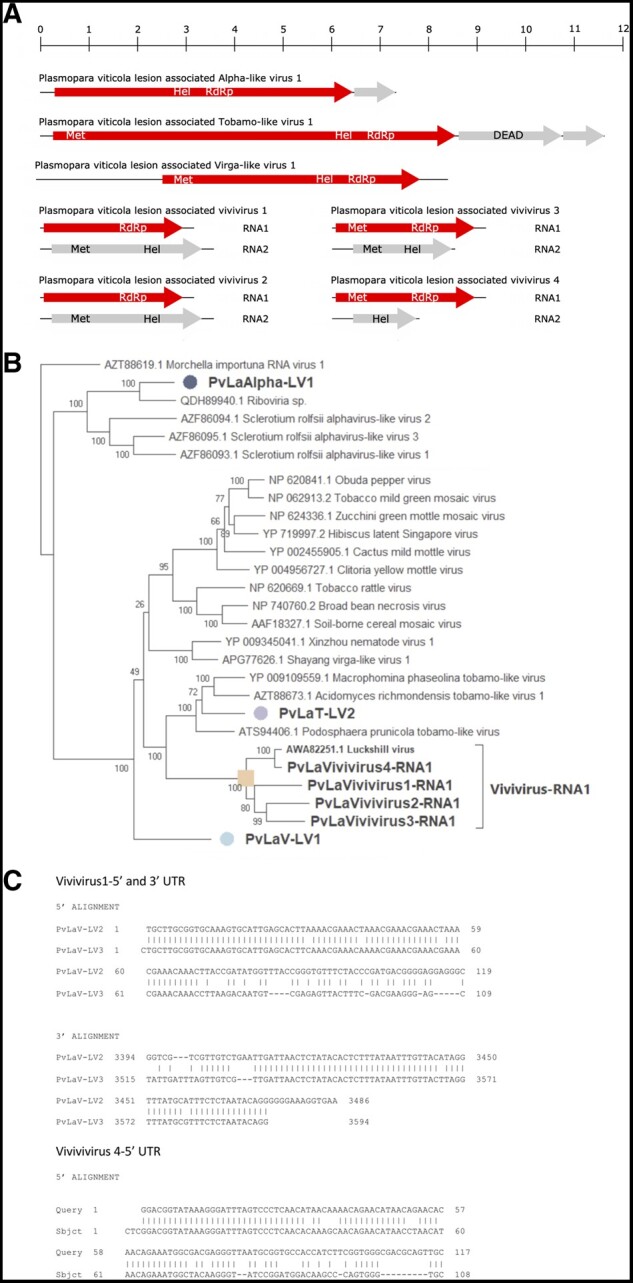
(A) Tobamo-virga-alpha genome organizations. Top ruler indicates the length in kb. (B) Tobamo-virga-alpha virus tree computed by IQ-TREE stochastic algorithm to infer phylogenetic trees by maximum likelihood. Model of substitution: Blosum64+F-I+G4. Consensus tree is constructed from 1,000 bootstrap trees. Log-likelihood of consensus tree: −120571.621. Purple label indicates the tobamovirus, orange labels indicate viviviruses in different sub-clades, light blue label indicates virga-like virus, and the blue one indicates the alphavirus. At nodes the percentage bootstrap values. (C) Sequence alignment of vivivirus 5′ and 3′ UTRs.
