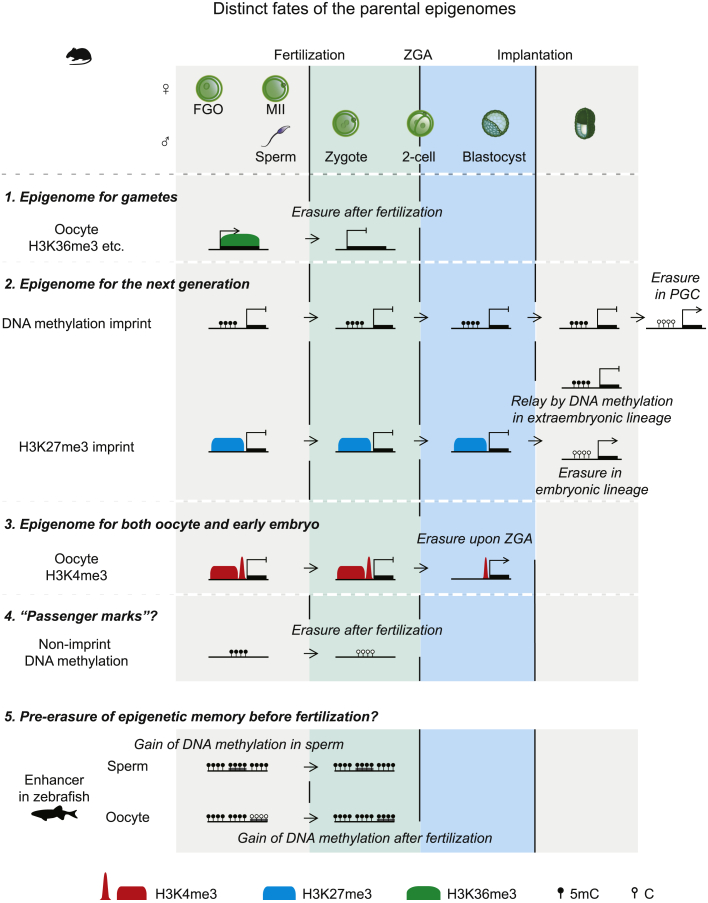
Figure 3.
Distinct Fates of the Parental Epigenomes
1. “Epigenome for gametes.” An example comes from oocyte H3K36me3, which is deposited in transcribed regions and plays critical functions in imprinting establishment and oocyte development. Oocyte H3K36me3 subsequently becomes possible “transcription fossils” when genome is silenced, and is removed after fertilization. 2. “Epigenome for the next generation.” Both DNA methylation- and maternal H3K27me3-mediated imprints are established in gametes and function in embryos. DNA methylation-mediated imprints can be faithfully maintained throughout development except in PGCs. The oocyte H3K27me3-mediated imprint can be inherited to pre-implantation embryos and is lost in post-implantation embryos, but is relayed by DNA methylation in extraembryonic lineages to maintain imprinting. 3. “Epigenome for both oocyte and early embryos.” Oocyte H3K4me3 is an example that plays roles in both mouse oocytes and embryos. In mouse FGOs, H3K4me3 promotes global genome silencing. In early embryos, H3K4me3 is required for establishing paternal LADs. 4. “Passenger marks?” Oocyte DNA methylation outside of ICRs is largely dispensable for oocyte development, raising the possibility that they may act as “passenger marks.” After fertilization, the majority of these DNA methylation marks undergoes global removal. 5. “Pre-erasure of parental epigenetic memory before fertilization.” The DNA methylation of enhancers often correlates with the loss of enhancer activities. While oocyte enhancers are progressively methylated after fertilization, sperm enhancers in zebrafish gain DNA methylation prior to fertilization, raising a possibility that the parental epigenetic memory (sperm enhancer) is pre-erased before fertilization. PGC, primordial germ cells.