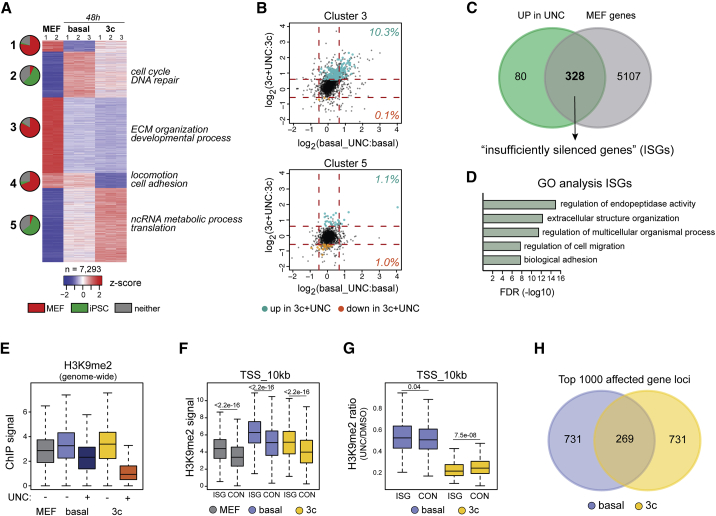
Figure 3.
Molecular Consequences of EHMT Inhibition during Early Reprogramming Stages
(A) Unsupervised k-means clustering of genes differentially expressed between MEFs and cells expressing OKSM for 2 days under either basal or 3c conditions (adjusted p < 0.05; fold change [FC] > 2). Pie charts indicate the abundance of genes associated with either MEFs or iPSCs (adjusted p < 0.05; FC > 2) or neither cell type in the respective cluster. Select GO categories (false discovery rate [FDR] < 0.05) associated with identified clusters are highlighted.
(B) Effects of EHMT inhibition (UNC) on the expression levels of genes associated with cluster 3 (top) and cluster 5 (bottom) identified in (A) during both basal (x axis) and 3c enhanced (y axis) reprogramming. Transcripts with significantly changed abundance during 3c enhanced reprogramming (adjusted p < 0.05; FC > 1.5) are highlighted in green (failed downregulation) and orange (failed upregulation), respectively. Percentage of affected cluster-specific genes is shown.
(C) Venn diagram showing overlap between genes upregulated during 3c enhanced reprogramming in the presence of UNC0638 and genes significantly more highly expressed in MEFs than in iPSCs.
(D) GO terms associated with MEF-associated genes that are inefficiently silenced during 3c enhanced reprogramming in the presence of UNC0638 (ISGs).
(E) H3K9me2 ChIP signal across 2.5-kb tiling intervals across the mouse genome. Average values from two separate chromatin precipitations are shown for each of the indicated conditions.
(F) H3K9me2 ChIP signal over the 10 kb immediately upstream of the TSS in ISGs in the presence of UNC0638 during 3c reprogramming and 2,664 control MEF-associated genes that are efficiently silenced (CON). n = 2 independent experiments.
(G) Ratio of H3K9me2 signal in the presence and absence of UNC0638 at the TSS region of ISGs and control genes during basal and 3c reprogramming. Indicated p values in (F and G) were calculated with Wilcoxon rank-sum test. n = 2 independent experiments.
(H) Overlap of the 1,000 protein-coding gene loci with the highest degree of loss of H3K9me2 signal upon EHMT inhibition during basal and 3c reprogramming.