Figure 4.
MeCP2 Targeting to Chromocenters Is Partly Dependent on the MajSat-fw Transcript
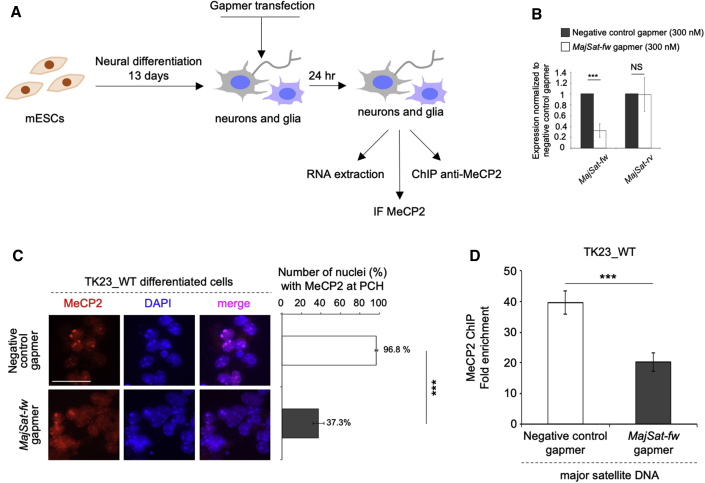
(A) Scheme of the experimental procedure.
(B) Strand-specific RT-qPCR analysis of MajSat-fw and MajSat-rv transcripts in mESC-derived TK23_WT neurons transfected with 300 nM negative control or MajSat-fw gapmers. Data are normalized to Gapdh and are means ± SD of transcript levels relative to cells transfected with negative control gapmer, from three biological replicates, with each amplified twice. ∗∗∗p < 0.001; NS, not significant (one-tailed Student’s t test).
(C) Left: Representative immunofluorescence of MeCP2 in mESC-derived TK23_WT neurons transfected with negative control or MajSat-fw gapmers. Chromocenters were stained with DAPI. Scale bar, 25 μm. Right: Quantification of MeCP2 enrichment at PCH following transfection of negative control or MajSat-fw gapmers. Data are means ± SD, with ≥100 cells analyzed per condition, from four independent experiments. ∗∗∗p < 0.001 (one-tailed Student’s t test).
(D) ChIP–qPCR of MeCP2 at MajSat DNA in mESC-derived TK23_WT neurons transfected with negative control or MajSat-fw gapmers. Data are means ± SD of four independent qPCR replicates from two ChIP experiments. ∗∗∗p < 0.001; NS, not significant (one-tailed Student’s t test). See also Figure S3.