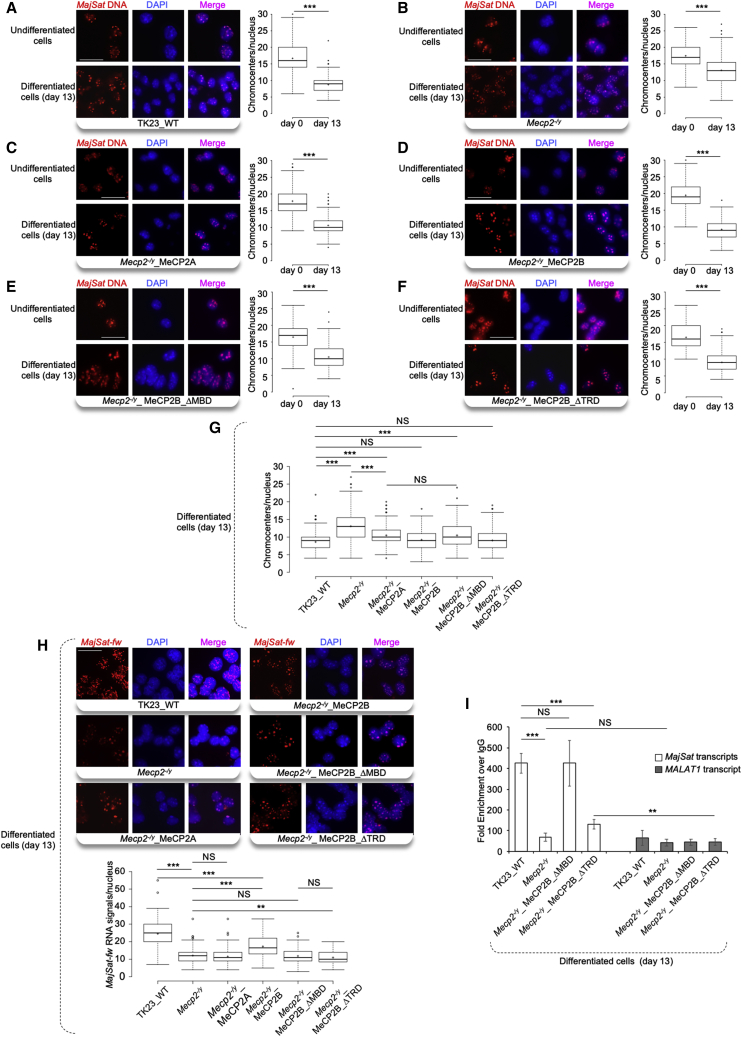
Figure 6.
MeCP2B and Its MBD Are the Major Players for Chromocenter Clustering and Both MBD and TRD Contribute to MajSat-fw Transcript Targeting to PCH
(A–F) Left: Representative images of i3D-DNA FISH in TK23_WT, Mecp2−/y, Mecp2−/y_MeCP2A, Mecp2−/y_MeCP2B, Mecp2−/y_MeCP2B_ΔMBD, and Mecp2−/y_MeCP2B_ΔTRD undifferentiated cells (day 0) and terminally differentiated neurons (day 13) using the MajSat-fw LNA probe. Chromocenters were stained with DAPI. Scale bar, 25 μm. Right: Quantification of chromocenters in three-dimensional space of each nucleus in undifferentiated mESCs and terminally differentiated neurons.
(G) Direct comparisons of the data reported in (A–F) for terminally differentiated neurons.
(H) Top: Representative images of i3D-RNA FISH in terminally differentiated TK23_WT, Mecp2−/y, Mecp2−/y_MeCP2A, Mecp2−/y_MeCP2B, Mecp2−/y_MeCP2B_ΔMBD, and Mecp2−/y_MeCP2B_ΔTRD neurons, using the MajSat-fw LNA probe. Chromocenters were stained with DAPI. Scale bar, 25 μm. Bottom: Quantification of MajSat-fw RNA signals in three-dimensional space of each nucleus in terminally differentiated neurons. Data for TK23_WT and Mecp2−/y neurons are also reported in Figure 2C. For (A–H), data are shown as box and whisker plots with 200 nuclei analyzed per condition, from two replicate slides. ∗∗p < 0.01; ∗∗∗p < 0.001; NS, not significant (two-sample Kolmogorov-Smirnov test, without (A–F) and with (G, H) Benjamini–Hochberg correction).
(I) RNA immunoprecipitation of MajSat transcripts in mESC-derived Mecp2−/y_MeCP2B_ΔMBD, Mecp2−/y_MeCP2B_ΔTRD, TK23_WT (positive control), and Mecp2−/y (negative control) neurons, using an anti-MeCP2 antibody. Data are means ± SD of three independent qPCR replicates from one representative RIP experiment. Additional biological replicates yielded similar results. ∗∗∗p < 0.001; ∗∗p < 0.01; NS, not significant (one-tailed Student’s t test). See also Figures S4–S6.